Abstract
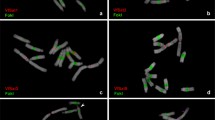
Giemsa C-banded idiograms that allow the identification of all chromosomes have been prepared for Allium cepa, Ornithogalum virens, and Secale cereale. An analysis of A. cepa DNA has determined that: (1) It has the lowest GC content so far reported for an angiosperm (∼32%). (2) It appears to have no satellite DNA detectable by CsCl or Cs2SO4-Ag+ density gradient centrifugation. (3) Aside from fold back DNA and unreactable fragments, a C0t curve indicates that most of the DNA can be adequately described as two major middle repetitive components (Fractions I and II) and a single copy component (Fraction III). And (4) most of the repeated DNA sequences are involved in a “short period” interspersion pattern with single copy and other repetitive sequences. In situ hybridization of tritiated cRNAs to fold back, long repeated, and Fraction I DNA from A. cepa to squash preparations of chromosomes and nuclei from A. cepa, O. virens, and S. cereale root tips indicates: (1) Sequences complementary to fold back DNA are scattered throughout the genome of A. cepa except for telomeric heterochromatin and nucleolus organizers while they are not detectable in the genomes of O. virens or S. cereale. (2) Although long repeated sequences are scattered throughout the genome of A. cepa, they are concentrated to some extent in telomeric heterochromatin and nucleolus organizers (NOs). Sequences complementary to long repeats of A. cepa occur primarily in chromosome three of O. virens while these sequences are more common in the genome of more distantly related S. cereale. (3) Fraction I DNA is scattered throughout the genome of A. cepa while it is hardly detectable in the genomes of O. virens and S. cereale. These results are discussed in regard to the evolutionary conservation and function of repeated DNA sequences.
Similar content being viewed by others
References
Bachmann, K., Price, H.: Repetitive DNA in Cichorieae (Compositae). Chromosoma (Berl.) 61, 267–275 (1977)
Bennett, M.: Nuclear DNA content and minimum generation time in herbaceous plants. Proc. roy. Soc. Lond., B, 181, 109–135 (1972)
Biswas, S., Sarkar, A.: Deoxyribonucleic acid base composition of some angiosperms and its significance. Phytochemistry 9, 2425–2430 (1970)
Britten, R., Graham, D., Eden, F., Painchaud, D., Davidson, E.: Evolutionary divergence and length of repetitive sequences in sea urchin DNA. J. molec. Evol. 9, 1–23 (1976)
Britten, R., Graham, D., Neufeld, B.: Analysis of repeating DNA sequences by reassociation. Methods in enzymology 29 E, 363–418 (1974)
Britten, R., Kohne, D.: Nucleotide sequence repetition in DNA. Carnegie Inst. Wash. Year Book 65, 78–106 (1966)
Britten, R., Kohne, D.: Repeated sequences in DNA. Science 161, 529–540 (1968)
Crain, W., Davidson, E., Britten, R.: Contrasting patterns of DNA sequence arrangement in Apis mellifera (honeybee) and Musca domestica (housefly). Chromosoma (Berl.) 59, 1–12 (1976)
Davidson, E., Graham, D., Neufield, B., Chamberlin, M., Amenson, C., Hough, B., Britten, R.: Arrangement and characterization of repetitive sequence elements in animal DNAs. Cold. Spr. Harb. Symp. quant. Biol. 38, 295–301 (1974)
Davidson, E., Hough, B., Amenson, C., Britten, R.: General interspersion of repetitive with non-repetitive sequence elements in the DNA of Xenopus. J. molec. Biol. 77, 1–23 (1973)
Deumling, B., Sinclair, J., Timmis, J., Ingle, J.: Demonstration of satellite DNA components in several plant species with the Ag+-Cs2SO4 gradient technique. Cytobiologie 13, 224–232 (1976)
Dott, P., Chuang, C., Saunders, G.: Inverted repetitive sequences in the human genome. Biochemistry 15, 4120–4125 (1976)
Doyle, J.: Fossil evidence on early evolution of the monocotyledons. Quart. Rev. Biol. 48, 399–413 (1973)
Eden, F., Graham, D., Davidson, E., Britten, R.: Exploration of long and short repetitive sequence relationships in the sea urchin genome. Nucleic Acids Res. 4, 1553–1567 (1977)
El-Gadi, A., Elkington, T.: Comparison of the Giemsa C-Band karyotypes and the relationships of Allium cepa, A. fistulosum, and A. galanthum. Chromosoma (Berl.) 51, 19–23 (1975)
Elgin, S., Weintraub, H.: Chromosomal proteins and chromatin structure. Ann. Rev. Biochem. 44, 725–774 (1975)
Fiskesjo, C.: Two types of constitutive heterochromatin made visible in Allium by a rapid C-banding method. Hereditas (Lund) 78, 153–156 (1974)
Flavell, R., Bennett, M., Smith, J., Smith, D.: Genome size and the proportion of repeated nucleotide sequence DNA in plants. Biochem. Genet. 12, 257–269 (1974)
Flavell, R., Rimpau, J., Smith, D.: Repeated sequence DNA relationships in four cereal genomes. Chromosoma (Berl.) 63, 205–222 (1977)
Flavell, R., Smith, D.: Nucleotide sequence organisation in the wheat genome. Heredity 37, 231–252 (1976)
Galau, G., Chamberlin, M., Hough, B., Britten, R., Davidson, E.: In: Molecular studies of biological evolution (F. Ayala, ed.), p. 200. Sunderland, Mass.: Sinauer Press 1976
Gall, J., Pardue, M.: Nucleic acid hybridization in cytological preparations. Methods in enzymology 21 D, 470–480 (1971)
Ginelli, E., Di Lernia, R., Corneo, G.: The organization of DNA sequences in the mouse genome. Chromosoma (Berl.) 61, 215–226 (1977)
Godin, D., Stack, S.: Homologous and non-homologous chromosome associations by interchromosomal chromatin connectives in Ornithogalum virens. Chromosoma (Berl.) 57, 309–318 (1976)
Hadlaczky, G., Koczka, K.: C-banding karyotype of rye from hexaploid triticale. Cereal Res. Commun. 2, 193–200 (1974)
Henderson, A., Warburton, D., Atwood, K.: Location of ribosomal DNA in the human chromosome complement. Proc. nat. Acad. Sci. (Wash.) 69, 3394–3398 (1972)
Huguet, T., Jouanin, L.: The heterogeneity of wheat DNA. Biochim. biophys. Acta (Amst.) 262, 431–440 (1972)
Ingle, H., Pearson, G., Sinclair, J.: Species distribution and properties of nuclear satellite DNA in higher plants. Nature (Lond.) New Biol. 242, 192–197 (1973)
Karavanov, A., Iordanskii, A.: The genome structure of Allium cepa L. and Allium fistulosum L. Mol. Biol. 7, 366–371 (1973)
Kiper, M., Herzfeld, F.: DNA sequence organization in the genome of Petroselinum sativum (Umbelliferae). Chromosoma (Berl.) 65, 335–351 (1978)
Kirk, J., Rees, H., Evans, G.: Base composition of nuclear DNA within the genus Allium. Heredity 25, 507–512 (1970)
Klein, W., Murphy, W., Attardi, G., Britten, R., Davidson, E.: Distribution of repetitive and nonrepetitive sequence transcripts in HeLa mRNA. Proc. nat. Acad. Sci. (Wash.) 71, 1785–1789 (1974)
MacGregor, H., Jones, C.: Chromosomes, DNA sequences, and evolution in salamanders of the genus Aneides. Chromosoma (Berl.) 63, 1–9 (1977)
Mandel, M., Marmur, J.: Use of ultraviolet absorbance-temperature profile for determining the guanine plus cytosine content of DNA. Methods in enzymology 12B, 195–206 (1968)
Mandel, M., Schildkraut, C., Marmur, J.: Use of CsCl density gradient analysis for determining the guanine plus cytosine content of DNA. Methods in enzymology 12B, 184–195 (1968)
Maniatis, T., Jeffrey, A., Kleid, D.: Nucleotide sequence of the rightward operator of phage. Proc. nat. Acad. Sci. (Wash.) 72, 1184–1188 (1975)
Marmur, J.: A procedure for isolation of deoxyribonucleic acid from micro-organisms. J. molec. Biol. 3, 208–218 (1961)
Melli, M., Whitfield, C., Rao, K., Richardson, M., Bishop, J.: DNA-RNA hybridization in vast DNA excess. Nature (Lond.) New Biol. 231, 8–12 (1971)
Mizuno, S., Andrews, C., MacGregor, H.: Interspecific “common” repetitive sequences in salamanders of the Genus Plethodon. Chromosoma (Berl.) 58, 1–31 (1976)
Noll, H.: Polysomes: Analysis of structure and function. In: Techniques in protein biosynthesis (P. Campbell and J. Sargent, eds.), p. 107–179. New York: Academic Press 1969
Ohno, S.: Ancient linkage groups and frozen accidents. Nature (Lond.) 244, 259–262 (1973)
Pardue, M., Gall, J.: Nucleic acid hybridization to the DNA of cytological preparations. In: Methods in enzymology 10, 1–16 (1975)
Pearson, W., Davidson, E., Britten, R.: A program for least squares analysis of reassociation and hybridization data. Nucleic Acids Res. 4, 1727–1737 (1977)
Ranjekar, P., Lafountaine, J., Pallotta, D.: Characterization of repetitive DNA in rye (Secale cereale). Chromosoma (Berl.) 48, 427–440 (1974)
Ranjekar, P., Pallotta, D., Lafountaine, J.: Analysis of the genome of plants II. Characterization of repetitive DNA in barley (Hordeum vulgare) and wheat (Triticum aestivum). Biochim. biophys. Acta (Amst.) 425, 30–40 (1976)
Robertson, H., Dickson, E., Jelinek, W.: Determination of nucleotide sequences from doublestranded regions of HeLa cells nuclear RNA. J. molec. Biol. 111, 571–589 (1977)
Ryskov, A., Saunders, G., Farashyan, V., Georgiev, G.: Double helical regions in nuclear precursor of mRNA (pre-mRNA). Biochim. biophys. Acta (Amst.) 312, 152–164 (1973)
Sinclair, J., Brown, D.: Retention of common nucleotide sequences in the ribosomal deoxyribonucleic acid of eukaryotes and some of their physical characteristics. Biochemistry 10, 2761–2769 (1971)
Singh, L.: Purdom, I., Jones, K.: Effect of different denaturing agents on the detectability of specific DNA sequences of various base compositions by in situ hybridisation. Chromosoma (Berl.) 60, 377–389 (1977)
Smith, D., Flavell, R.: Nucleotide sequence organization in the rye genome. Biochim. biophys. Acta (Amst.) 474, 82–97 (1977)
Stack, S.: Differential Giemsa staining of kinetochores and nucleolus organizer heterochromatin in mitotic chromosomes of higher plants. Chromosoma (Berl.) 47, 361–378 (1974)
Studier, F.: Sedimentation studies of the size and shape of DNA. J. molec. Biol. 11, 373–390 (1965)
Vosa, C.: Heterochromatic patterns in Allium. I. The relationship between the species of the cepa group and its allies. Heredity 36, 383–392 (1976)
Walbot, V., Dure, L.: Developmental biochemistry of cotton seed embryogenesis and germination. VII. Characterization of the cotton genome. J. molec. Biol. 10, 503–536 (1976)
Wilson, D., Thomas, C.: Palindromes in chromosomes. J. molec. Biol. 84, 115–144 (1974)
Zimmerman, J., Goldberg, R.: DNA sequence organization in the genome of Nicotiana tabacum. Chromosoma (Berl.) 59, 227–252 (1977)
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Stack, S.M., Comings, D.E. The chromosomes and DNA of Allium cepa . Chromosoma 70, 161–181 (1979). https://doi.org/10.1007/BF00288404
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00288404