Summary
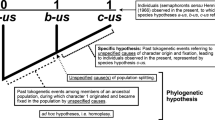
The accuracies and efficiencies of three different methods of making phylogenetic trees from gene frequency data were examined by using computer simulation. The methods examined are UPGMA, Farris' (1972) method, and Tateno et al.'s (1982) modified Farris method. In the computer simulation eight species (or populations) were assumed to evolve according to a given model tree, and the evolutionary changes of allele frequencies were followed by using the infinite-allele model. At the end of the simulated evolution five genetic distance measures (Nei's standard and minimum distances, Rogers' distance, Cavalli-Sforza's fλ, and the modified Cavalli-Sforza distance) were computed for all pairs of species, and the distance matrix obtained for each distance measure was used for reconstructing a phylogenetic tree. The phylogenetic tree obtained was then compared with the model tree. The results obtained indicate that in all tree-making methods examined the accuracies of both the topology and branch lengths of a reconstructed tree (rooted tree) are very low when the number of loci used is less than 20 but gradually increase with increasing number of loci. When the expected number of gene substitutions (M) for the shortest branch is 0.1 or more per locus and 30 or more loci are used, the topological error as measured by the distortion index (dT) is not great, but the probability of obtaining the correct topology (P) is less than 0.5 even with 60 loci. When M is as small as 0.004, P is substantially lower. In obtaining a good topology (small dT and high P) UPGMA and the modified Farris method generally show a better performance than the Farris method. The poor performance of the Farris method is observed even when Rogers' distance which obeys the triangle inequality is used. The main reason for this seems to be that the Farris method often gives overestimates of branch lengths. For estimating the expected branch lengths of the true tree UPGMA shows the best performance. For this purpose Nei's standard distance gives a better result than the others because of its linear relationship with the number of gene substitutions. Rogers' or Cavalli-Sforza's distance gives a phylogenetic tree in which the parts near the root are condensed and the other parts are elongated. It is recommended that more than 30 loci, including both polymorphic and monomorphic loci, be used for making phylogentic trees. The conclusions from this study seem to apply also to data on nucleotide differences obtained by the restriction enzyme techniques.
Similar content being viewed by others
References
Avise JC, Lansman RA, Shade RO (1979) The use of restriction endonucleases to measure mitochondrial DNA sequence relatedness in natural populations. I. Population structure and evolution in the genus Peromyscus. Genetics 92:279–295
Bhattacharrya A (1946) On a measure of divergence between two multinomial pupulations. Sankhya 7:401–406
Brown WM, George Jr. M, Wilson AC (1979) Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci 76:1967–1971
Cavalli-Sforza LL (1969) Human Diversity. Proc 12th Intl Cong Genet, Tokyo, Vol 3:405–416
Cavalli-Sforza LL, Edwards AWF (1967) Phylogenetic analysis: models and estimation procedures. Amer J Hum Gen 19: 233–257
Cavalli-Sforza LL, Piazza A (1975) Analysis of evolution: Evolutionary rates, independence and treeness. Theoret Pop Biol 8:127–165
Chakraborty R (1977) Estimation of time of divergence from phylogenetic studies. Can J Genet Cytol 19:217–223
Chakraborty R, Nei M (1977) Bottleneck effects on average heterozygosity and genetic distance with the stepwise mutation model. Evolution 31:347–356
Chakraborty R, Fuerst PA, Nei M (1977) A comparative study of genetic variation within and between populations under the neutral mutation hypothesis and the model of sequentially advantageous mutations. (Abstract) Genetics 86:s10–11
Farris JS (1972) Estimating phylogenetic trees from distance matrices. Amer Nat 106:645–668
Farris JS (1981) Distance data in phylogenetic analysis. In: Funk VA, Brooks DR (eds) Advances in cladistics. Proc. 1st Meeting of Willi Hennig Society, Publ. New York Botanical Garden, Bronx, NY, pp 1–23
Fitch WM, Margoliash E (1967) Construction of phylogenetic trees. Science 155:279–284
Gotoh O, Hayashi JI, Yonekawa H, Tagashira Y (1979) An improved method for estimating sequence divergence between related DNAs from changes in restriction endonuclease cleavage sites. J Mol Evol 14:301–310
Griffiths RC (1980) Lines of descent in the diffusion approximation of neutral Wright-Fisher models. Theoret Pop Biol 17:37–50
Griffiths RC, Li WH (1983) Simulating allele frequencies in a population and the genetic differentiation of populations under mutation pressure. Theoret Pop Biol (in press)
Kaplan N, Langley CH (1979) A new estimate of sequence divergence of mitochondrial DNA using restriction endonuclease mappings. J Mol Evol 13:295–304
Kidd KK, Cavalli-Sforza LL (1971) Number of characters examined and error in reconstruction of evolutionary trees. In: Hodson FR, Kendall DG, Tautu P (eds) Mathematics in the archaeological and historical sciences. Edinburgh University Press, Edinburgh, pp 335–346
Kimura M, Crow JF (1964) The number of alleles that can be maintained in a finite population. Genetics 49:725–738
Li WH (1976) Effect of migration on genetic distance. Amer Nat 110:841–847
Li WH, Nei M (1975) Drift variances of heterozygosity and genetic distance in transient states. Genet Res 25:229–248
Nei M (1972) Genetic distance between populations. Amer Nat 106:283–292
Nei M (1973) The theory and estimation of genetic distance. In: Morton NE (ed) Genetic structure of populations. University of Hawaii Press, Honolulu, pp 45–54
Nei M (1975) Molecular population genetics and evolution. North Holland, Amsterdam and New York
Nei M (1976) Mathematical models of speciation and genetic distance. In: Karlin S, Nevo E (eds) Population genetics and ecology, Academic Press, New York, pp 723–765
Nei M (1977) Standard error of immunological dating of evolutionary time. J Mol Evol 9:203–211
Nei M (1978a) The theory of genetic distance and evolution of human races. Japan J Hum Genet 23:341–369
Nei M (1978b) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci 76:5269–5273
Nei M, Roychoudhury AK (1974) Sampling variances of heterozygosity and genetic distance. Genetics 76:379–390
Nei M, Tateno Y (1975) Interlocus variation of genetic distance and the neutral mutation theory. Proc Natl Acad Sci 72: 2758–2760
Prager EM, Wilson AC (1978) Construction of phylogenetic trees for proteins and nucleic acids: comparison of alternative matrix methods. J Mol Evol 11:129–142
Robinson DF, Foulds LR (1981) Comparison of phylogenetic trees. Math Biosci 53:131–147
Rogers JS (1972) Measures of genetic similarity and genetic distance. Studies in Genetics VII (University of Texas Publ. No. 7213), pp 145–153
Sanghvi LD (1953) Comparison of genetical and morphological methods for a study of biological differences. Amer J Phys Anthrop 11:385–404
Sarich VM, Wilson AC (1967) Immunological time scale for hominid evolution. Science 158:1200–1203
Shah DM, Langley CH (1979) Inter-and intraspecific variation in restriction maps ofDrosophila mitochondrial DNAs. Nature 281:696–699
Sneath PHA, Sokal RR (1973) Numerical taxonomy. WH Freeman, San Francisco
Swofford DL (1981) On the utility of the distance Wagner procedure. In: Funk VA, Brooks DR (eds) Advances in cladistics. Proc. 1st Meeting of Willi Hennig Society, Publ. New York Botanical Garden, Bronx, NY, pp 25–43
Tateno Y (1982) Statistical examination of phylogenetic tree construction methods by computer simulation. In: Kimura M (ed) Molecular evolution, protein polymorphism and the neutral theory. Japan Scientific Societies Press, Tokyo/ Springer-Verlag, Berlin, pp 217–229
Tateno Y, Nei M, Tajima F (1982) Accuracy of estimated phylogenetic trees from molecular data. I. Distantly related species. J Mol Evol 18:387–404
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Nei, M., Tajima, F. & Tateno, Y. Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19, 153–170 (1983). https://doi.org/10.1007/BF02300753
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF02300753