Abstract
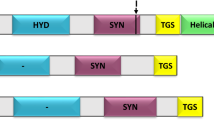
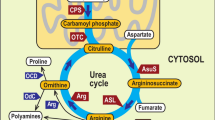
Tryptophan is an essential amino acid that, in eukaryotes, is synthesized either in the plastids of photoautotrophs or in the cytosol of fungi and oomycetes. Here we present an in silico analysis of the tryptophan biosynthetic pathway in stramenopiles, based on analysis of the genomes of the oomycetes Phytophthora sojae and P. ramorum and the diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum. Although the complete pathway is putatively located in the complex chloroplast of diatoms, only one of the involved enzymes, indole-3-glycerol phosphate synthase (InGPS), displays a possible cyanobacterial origin. On the other hand, in P. tricornutum this gene is fused with the cyanobacteria-derived hypothetical protein COG4398. Anthranilate synthase is also fused in diatoms. This fusion gene is almost certainly of bacterial origin, although the particular source of the gene cannot be resolved. All other diatom enzymes originate from the nucleus of the primary host (red alga) or secondary host (ancestor of chromalveolates). The entire pathway is of eukaryotic origin and cytosolic localization in oomycetes; however, one of the enzymes, anthranilate phosphoribosyl transferase, was likely transferred to the oomycete nucleus from the red algal nucleus during secondary endosymbiosis. This suggests possible retention of the complex plastid in the ancestor of stramenopiles and later loss of this organelle in oomycetes.









Similar content being viewed by others
References
Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105
Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, Brzezinski MA, Chaal BK, Chiovitti A, Davis AK, Demarest MS, Detter JC, Glavina T, Goodstein D, Hadi MZ, Hellsten U, Hildebrand M, Jenkins BD, Jurka J, Kapitonov VV, Kroger N, Lau WW, Lane TW, Larimer FW, Lippmeier JC, Lucas S, Medina M, Montsant A, Obornik M, Parker MS, Palenik B, Pazour GJ, Richardson PM, Rynearson TA, Saito MA, Schwartz DC, Thamatrakoln K, Valentin K, Vardi A, Wilkerson FP, Rokhsar DS (2004) The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306:79–86
Atkinson DE (1977) Cellular energy metabolism and its regulation. Academic Press, New York
Bae YM, Crawford IP (1990) The Rhizobium meliloti TrpE(G) gene is regulated by attenuation, and its product Anthranilate synthase is regulated by feedback inhibition. J Bacteriol 172(6):3318–3327
Bartholmes P, Böker H, Jaenicke R (1979) Purification of tryptophan synthase from Saccharomyces cerevisiae and partial activity of its nicked subunits. Eur J Biochem 102:167–172
Bodyl A (2005) Do plastid-related characters support the chromalveolate hypothesis? J Phycol 41:712–719
Bohlmann J, De Luca V, Eilert U, Martin W (1995) Purification and cDNA cloning of anthranilate synthase from Ruta graveolens: models of expression and properties of native recombinant enzymes. Plant J 7:491–501
Braus GH (1991) Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for regulation of a eukaryotic biosynthetic pathway. Microbiol Rev 55:349–370
Caliguiri MG, Bauerle R (1991) Subunit communication in the anthranilate synthase complex from Salmonella typhimurinum. Science 252:1845–1848
Cavalier-Smith T (1999) Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J Eukaryot Microbiol 46:347–366
Cavalier-Smith T (2002) Chloroplast evolution: secondary symbiogenesis and multiple losses. Curr Biol 12:R62–R64
Cheresh P, Harrison T, Fujioka H, Haldari K (2002) Targeting the malarial plastid via the parasitoforous vacuole. J Biol Chem 227:1265–1277
Crawford IP (1975) Gene rearrangements in the evolution of the tryptophan synthetic patway. Bacteriol Rev 39:87–120
Crawford IP (1989) Evolution of biosynthetic pathway: tryptophan paradigm. Annu Rev Microbiol 43:567–600
Creighton TE, Yanofsky C (1970) Chorismate to tryptophan (Escherichia coli)—anthranilate synthetase, PR transferase, PRA isomerase, InGP synthetase, tryptophan synthetase. Methods Enzymol 17:365–380
DeMoss JA, Wegman J (1965) An eznyme aggregate in tryptophan pathway of Neurospora crassa. Proc Natl Acad Sci USA 54:241–247
DeRocher A, Hagen CB, Froehlich JE, Feagin JE, Parsons M (2000) Analysis of targeting sequences demonstrates that trafficking to the Toxoplasma gondii plastid branches off the secretory system. J Cell Sci 113:3969–3977
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016
Falkowski PG, Katz ME, Knoll AH, Quigg A, Raven JA, Schofield O, Taylor FJR (2004) The evolution of modern eukaryotic phytoplankton. Science 305:354–360
Green JM, Nichols BP (1991) p-Aminobenzoate biosynthesis in Escherichia coli. J Biol Chem 266:12971–12975
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Harb OS, Chatterjee B, Fraunholz MJ, Crawford MJ, Nishi M, Roos DS (2004) Multiple functionally redundant signals mediate targeting to the apicoplast in the apicomplexan parasite Toxoplasma gondii. Euk Cell 3:663–674
Hasegawa M, Kishino K, Yano T (1985) Dating the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Hütter RP, Niederberger JA, DeMoss (1986) Tryptophan biosynthetic genes in eukaryotic microorganisms. Annu Rev Microbiol 40:55–77
Illingworth C, Mayer MJ, Elliott K, Hanfrey C, Walton NJ, Michael AJ (2003) The diverse bacterial origins of the Arabidopsis polyamine biosynthetic pathway. FEBS Lett 549:26–30
Ishida K (2005) Protein targeting into plastids: a key to understanding the symbiogenetic acquisition of plastids. J Plant Res 118:237–245
Kilian O, Kroth PG (2005) Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J 41:175–183
Kroth PG (2002) Protein transport into secondary plastids and the evolution of primary and secondary plastids. Int Rev Cyt Cell Biol 221:191–255
Kummerfeld SK, Teichmann SA (2005) Relative rates of gene fusion and fission in multi-domain proteins. Trends Genet 21:25–30
Lockhart PJ, Steel MA, Hendy MD, Penny D (1994) Recovering evolutionary trees under a more realistic model of sequence evolution. Mol Biol Evol 11:605–612
Mackenzie SA (2005) Plant organellar protein targeting: a traffic plan still under construction. Trends Cell Biol 15(10):548–554
Maheswari U, Montsant A, Goll J, Krishnasamy S, Rajyashri KR, Patell VM, Bowler C (2005) The Diatom EST Database. Nucleic Acids Res 33:D344–D347
Matchett WH, DeMoss JA (1975) The subunit structure of tryptophan synthase from Neurospora crassa. J Biol Chem 250:2941–2946
Matern U. (19940 Dianthus species (Carnations): In vitro culture and biosynthesis of dianthalexin and other secondary metabolites. In: Bajaj YPS (ed) Biotechnology in agriculture and forestry. Vol 28. Medicinal and aromatic plants VII. Springer-Verlag, Heidelberg, pp 170–184
Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Nishida K, Yoshida Y, Nishimura Y, Nakao S, Kobayashi T, Momoyama Y, Higashiyama T, Minoda A, Sano M, Nomoto H, Oishi K, Hayashi H, Ohta F, Nishizaka S, Haga S, Miura S, Morishita T, Kabeya Y, Terasawa K, Suzuki Y, Ishii Y, Asakawa S, Takano H, Ohta N, Kuroiwa H, Tanaka K, Shimizu N, Sugano S, Sato N, Nozaki H, Ogasawara N, Kohara Y, Kuroiwa T (2004) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428:653–657
McFadden GI (1999) Plastids and protein targeting. J Euk Microbiol 46:339–346
McFadden GI (2001) Primary and secondary endosymbiosis and the origin of plastids. J Phycol 37:951–959
Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10:1–6
Nielsen H, Brunak S, von Heijne G (1999) Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng 12:3–9
Obornik M, Green BR (2005) Mosaic origin of the heme biosynthesis pathway in photosynthetic eukaryotes. Mol Biol Evol 22:2343–2353
Ohta N, Sato N, Kawano S, Kuroiwa T (1993) The trpA gene on the plastid genome of Cyanidium caldarium strain RK-1. Curr Genet 25:357–361
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Prantl F, Strasser A, Aebi M, Furter R, Niederberger P (1985) Purification and characterization of the indole-3-glycerolphosphate synthase/anthranilate synthase complex of Saccharomyces cerevisiae. Eur J Biochem 146:95–100
Radwanski ER, Last RL (1995) Tryptophan biosynthesis and metabolism: biochemical and molecular genetics. Plant Cell 7:921–934
Richards TA, Dacks JB, Campbell SA, Blanchard JL, Foster PG, McLeod R, Roberts CW (2006) Evolutionary origins of the eukaryotic shikimate pathway: gene fusions, horizontal gene transfer, and endosymbiotic replacements. Eukaryot Cell 5:1517–1531
Siddall ME, Whiting MF (1999) Long-branch abstractions. Cladistics 15:9–24
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 15:4876–4882
Tyler BM, Tripathy S, Zhang XM, Dehal P, Jiang RHY, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL, Chapman J, Damasceno CMB, Dorrance AE, Dou DL, Dickerman AW, Dubchak IL, Garbelotto M, Gijzen M, Gordon SG, Govers F, Grunwald NJ, Huang W, Ivors KL, Jones RW, Kamoun S, Krampis K, Lamour KH, Lee MK, McDonald WH, Medina M, Meijer HJG, Nordberg EK, Maclean DJ, Ospina-Giraldo MD, Morris PF, Phuntumart V, Putnam NH, Rash S, Rose JKC, Sakihama Y, Salamov AA, Savidor A, Scheuring CF, Smith BM, Sobral BWS, Terry A, Torto-Alalibo TA, Win J, Xu ZY, Zhang HB, Grigoriev IV, Rokhsar DS, Boore JL (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313:1261–1266
Xie G, Forst C, Bonner C, Jensen RA (2002) Significance of two distinct types of tryptophan synthase beta chain in Bacteria, Archaea and higher plants. Genome Biol 3:0004.1–0004.13
Yanofsky C, Platt T, Crawford IP, Nichols BP, Christie GE, Horowitz H, Vancleemput M, Wu AM (1981) The complete nucleotide sequence of the tryptophan operon of Escherichia coli. Nucleic Acids Res 9:6647–6668
Yoon HS, Hacket JD, Pinto G, Bhattacharya D (2004) Molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol 21:809–818
Zhao J, Last RL (1995) Immunological characterization and chloroplast import of the tryptophan biosynthetic enzymes of the flowering plant Arabidopsis thaliana. J Biol Chem 270:6081–6087
Acknowledgments
This work was supported by the Grant Agency of the Academy of Sciences of the Czech Republic, Project No. IAA500220502 and Research Plan No. z60220518, and Ministry of Education of the Czech Republic, project no: 6007665801. We thank A. Lilley for critical reading of the manuscript and Beverley R. Green for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Electronic supplementary material
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Jiroutová, K., Horák, A., Bowler, C. et al. Tryptophan Biosynthesis in Stramenopiles: Eukaryotic Winners in the Diatom Complex Chloroplast. J Mol Evol 65, 496–511 (2007). https://doi.org/10.1007/s00239-007-9022-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-007-9022-z