Abstract
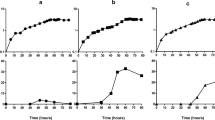
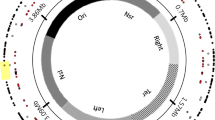
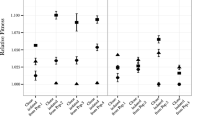
Insertion sequence (IS) elements are present in almost all bacterial genomes and are mobile enough to provide genomic tools to differentiate closely related isolates. They can be used to estimate genetic diversity and identify fitness-enhancing mutations during evolution experiments. Here, we determined the genomic distribution of eight IS elements in 120 genomes sampled from Escherichia coli populations that evolved in glucose- and phosphate-limited chemostats by comparison to the ancestral pattern. No significant differential transposition of the various IS types was detected across the environments. The phylogenies revealed substantial diversity amongst clones sampled from each chemostat, consistent with the phenotypic diversity within populations. Two IS-related changes were common to independent chemostats, suggesting parallel evolution. One of them corresponded to insertions of IS1 elements within rpoS encoding the master regulator of stress conditions. The other parallel event was an IS5-dependent deletion including mutY involved in DNA repair, thereby providing the molecular mechanism of generation of mutator clones in these evolving populations. These deletions occurred in different co-existing genotypes within single populations and were of various sizes. Moreover, differential locations of IS elements combined with their transpositional activity provided evolved clones with different phenotypic landscapes. Therefore, IS elements strongly influenced the evolutionary processes in continuous E. coli cultures by providing a way to modify both the global regulatory network and the mutation rates of evolving cells.



Similar content being viewed by others
References
Alokam S, Liu SL, Said K, Sanderson KE (2002) Inversions over the terminus region in Salmonella and Escherichia coli: IS200s as the sites of homologous recombination inverting the chromosome of Salmonella enterica serovar typhi. J Bacteriol 184:6190–6197
Atwood KC, Schneider LK, Ryan FJ (1951) Periodic selection in Escherichia coli. Proc Natl Acad Sci USA 37:146–155
Bartosik D, Putyrski M, Dziewit L, Malewska E, Szymanik M, Jagiello E, Lukasik J, Baj J (2008) Transposable modules generated by a single copy of insertion sequence ISPme1 and their influence on structure and evolution of natural plasmids of Paracoccus methylutens DM12. J Bacteriol 190:3306–3313
Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V et al (1997) The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462
Blot M (1994) Transposable elements and adaptation of host bacteria. Genetica 93:5–12
Chao L, Vargas C, Spear BB, Cox EC (1983) Transposable elements as mutator genes in evolution. Nature 303:633–635
Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220
Chou H-H, Berthet J, Marx CJ (2009) Fast growth increases the selective advantage of a mutation arising recurrently during evolution under metal limitation. PLoS Genet 5(9):e1000652
Cooper VS, Schneider D, Blot M, Lenski RE (2001) Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. J Bacteriol 183:2834–2841
Cordaux R, Pichon S, Ling A, Pérez P, Delaunay C, Vavre F, Bouchon D, Grève P (2008) Intense transpositional activity of insertion sequences in an ancient obligate endosymbiont. Mol Biol Evol 25:1889–1896
de Visser JAGM, Akkermans ADL, Hoekstra RF, de Vos WM (2004) Insertion-sequence-mediated mutations isolated during adaptation to growth and starvation in Lactococcus lactis. Genetics 168:1145–1157
Dekel E, Alon U (2005) Optimality and evolutionary tuning of the expression level of a protein. Nature 436:588–592
Drevinek P, Baldwin A, Lindenburg L, Joshi LT, Marchbank A, Vosahlikova S, Dowson CG, Mahenthiralingam E (2010) Oxidative stress of Burkholderia cenocepacia induces insertion sequence-mediated genomic rearrangements that interfere with macrorestriction-based genotyping. J Clin Microbiol 48:34–40
Eichenbaum Z, Livneh Z (1998) UV light induces IS10 transposition in Escherichia coli. Genetics 149:1173–1181
Felsenstein J (1989) PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5:164–166
Ferenci T (1999) Regulation by nutrient limitation. Curr Opin Microbiol 2:208–213
Ferenci T (2008a) Bacterial physiology, regulation and mutational adaptation in a chemostat environment. Adv Microb Physiol 53:169–229
Ferenci T (2008b) The spread of a beneficial mutation in experimental bacterial populations: the influence of the environment and genotype on the fixation of rpoS mutations. Heredity 100:446–452
Ferenci T, Zhou Z, Betteridge T, Ren Y, Liu Y, Feng L, Reeves PR, Wang L (2009) Genomic sequencing reveals regulatory mutations and recombinational events in the widely used MC4100 lineage of Escherichia coli K-12. J Bacteriol 191:4025–4029
Futuyama DJ (1986) Evolutionary biology, 2nd edn. Sinauer, Sunderland
Gerrish PJ, Lenski RE (1998) The fate of competing beneficial mutations in an asexual population. Genetica 102:127–144
Golubov A, Gierczynski R, Heesemann J, Rakin A (2005) A novel insertion sequence element, ISYen2, as an epidemiological marker for weakly pathogenic bioserotypes of Yersinia enterocolitica. Int J Med Microbiol 295:213–226
Gregory ST, Dahlberg AE (2008) Transposition of an insertion sequence, ISTth7, in the genome of the extreme thermophile Thermus thermophilus HB8. FEMS Microbiol Lett 289:187–192
Hall BG (1999) Spectra of spontaneous growth-dependent and adaptive mutations at ebgR. J Bacteriol 181:1149–1155
Hall BG, Parker LL, Betts PW, DuBose RF, Sawyer SA, Hartl DL (1989) IS103, a new insertion element in Escherichia coli: characterization and distribution in natural populations. Genetics 121:423–431
Hengge-Aronis R (2002) Recent insights into the general stress response regulatory network in Escherichia coli. J Mol Microbiol Biotechnol 4:341–346
Jeong H, Barbe V, Lee CH, Vallenet D, Yu DS, Choi SH et al (2009) Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J Mol Biol 394:644–652
Kaleta P, O’Callaghan J, Fitzgerald GF, Beresford TP, Ross RP (2010) Crucial role for insertion sequence elements in Lactobacillus helveticus evolution as revealed by interstrain genomic comparison. Appl Environ Microbiol 76:212–220
Kichenaradja P, Siguier P, Pérochon J, Chandler M (2010) ISbrowser: an extension of ISfinder for visualizing insertion sequences in prokaryotic genomes. Nucleic Acids Res 38(Database issue):D62–D68
Kitamura K, Torii Y, Matsuoka C, Yamamoto K (1995) DNA sequence changes in mutations in the tonB gene on the chromosome of Escherichia coli K-12: Insertion elements dominate the mutational spectra. Jpn J Genet 70:35–46
LeClerc JE, Li B, Payne WL, Cebula T (1996) High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208–1211
Maharjan R, Seeto S, Notley-McRobb L, Ferenci T (2006) Clonal adaptive radiation in a constant environment. Science 313:514–517
Maharjan RP, Seeto S, Ferenci T (2007) Divergence and redundancy of transport and metabolic rate-yield strategies in a single Escherichia coli population. J Bacteriol 189:2350–2358
Maharjan R, Zhou Z, Ren Y, Li Y, Gaffé J, Schneider D, McKenzie C, Reeves PR, Ferenci T, Wang L (2010) Genomic identification of a novel mutation in hfq that provides multiple benefits in evolving glucose-limited populations of Escherichia coli. J Bacteriol 192:4517–4521
Mahillon J, Chandler M (1998) Insertion sequences. Microbiol Mol Biol Rev 62:725–774
McClintock B (1965) The control of gene expression in maize. Brookhaven Symp Biol 18:162–184
Miller J (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, New York
Miller JH (1996) Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu Rev Microbiol 50:625–643
Naas T, Blot M, Fitch WM, Arber W (1994) Insertion sequence-related genetic variation in resting Escherichia coli K-12. Genetics 136:721–730
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Notley-McRobb L, Ferenci T (1999) The generation of multiple co-existing mal-regulatory mutations through polygenic evolution in glucose-limited populations of Escherichia coli. Environ Microbiol 1:45–52
Notley-McRobb L, Ferenci T (2000) Experimental analysis of molecular events during mutational periodic selections in bacterial evolution. Genetics 156:1493–1501
Notley-McRobb L, Pinto R, Seeto S, Ferenci T (2002) Regulation of mutY and nature of mutator mutations in Escherichia coli populations under nutrient limitation. J Bacteriol 184:739–745
Notley-McRobb L, Seeto S, Ferenci T (2003) The influence of cellular physiology on the initiation of mutational pathways in Escherichia coli populations. Proc R Soc Lond B 270:843–848
Papadopoulos D, Schneider D, Meier-Eiss J, Arber W, Lenski RE, Blot M (1999) Genomic evolution during a 10, 000-generation experiment with bacteria. Proc Natl Acad Sci USA 96:3807–3812
Pasternak C, Ton-Hoang B, Coste G, Bailone A, Chandler M, Sommer S (2010) Irradiation-induced Deinococcus radiodurans genome fragmentation triggers transposition of a single resident insertion sequence. PLoS Genet 6(1):e1000799
Pellicer MT, Badia J, Aguilar J, Baldoma L (1996) glc locus of Escherichia coli: characterization of genes encoding the subunits of glycolate oxidase and the glc regulator protein. J Bacteriol 178:2051–2059
Pelosi L, Kühn L, Guetta D, Garin J, Geiselmann J, Lenski RE, Schneider D (2006) Parallel changes in global protein profiles during long-term experimental evolution in Escherichia coli. Genetics 173:1851–1869
Peterson CN, Mandel MJ, Silhavy TJ (2005) Escherichia coli starvation diets: essential nutrients weigh in distinctly. J Bacteriol 187:7549–7553
Philippe N, Pelosi L, Lenski RE, Schneider D (2009) Evolution of penicillin-binding protein 2 concentration and cell shape during a long-term experiment with Escherichia coli. J Bacteriol 191:909–921
Reif HJ, Saedler H (1975) IS1 is involved in deletion formation in the gal region of E. coli K-12. Mol Gen Genet 137:17–28
Reynolds AE, Felton J, Wright A (1981) Insertion of DNA activates the cryptic bgl operon in Escherichia coli K-12. Nature 293:625–629
Riehle MM, Bennett AF, Long AD (2001) Genetic architecture of thermal adaptation in Escherichia coli. Proc Natl Acad Sci USA 98:525–530
Rodriguez H, Snow ET, Bhat U, Loechler EL (1992) An Escherichia coli plasmid-based, mutational system in which supF mutants are selectable: insertion elements dominate the spontaneous spectra. Mutat Res 270:219–231
Rohmer L, Fong C, Abmayr S, Wasnick M, Larson Freeman TJ, Radey M et al (2007) Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol 8:R102
Saedler H, Reif HJ, Hu S, Davidson N (1974) IS2, a genetic element for turn-off and turn-on of gene activity in Escherichia coli. Mol Gen Genet 132:265–289
Saitou N, Nei M (1987) The Neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sawyer SA, Dykhuisen DE, DuBose RF, Green L, Mutangadura-Mhlanga T, Wolczyk DF, Hartl DL (1987) Distribution and abundance of insertion sequences among natural isolates of Escherichia coli. Genetics 115:51–63
Schneider D, Duperchy E, Coursange E, Lenski RE, Blot M (2000) Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics 156:477–488
Schneider D, Duperchy E, Depeyrot J, Coursange E, Lenski RE, Blot M (2002) Genomic comparisons among Escherichia coli strains B, K-12, and O157:H7 using IS elements as molecular markers. BMC Microbiol 2:18
Siguier P, Filée J, Chandler M (2006) Insertion sequences in prokaryotic genomes. Curr Opin Microbiol 9:526–531
Sleight SC, Orlic C, Schneider D, Lenski RE (2008) Genetic basis of evolutionary adaptation by Escherichia coli to stressful cycles of freezing, thawing and growth. Genetics 180:431–443
Sniegowski PD, Gerrish PJ, Lenski RE (1997) Evolution of high mutation rates in experimental populations of Escherichia coli. Nature 387:703–705
Song H, Hwang J, Yi H, Ulrich RL, Yu Y, Nierman WC, Kim HS (2010) The early stage of bacterial genome-reductive evolution in the host. PLoS Pathog 6(5):e1000922
Stoebel DM, Dorman CJ (2010) The effect of mobile element IS10 on experimental regulatory evolution in Escherichia coli. Mol Biol Evol 27:2105–2112
Sun X, Dennis JJ (2009) A novel insertion sequence derepresses efflux pump expression and preadapts Pseudomonas putida S12 for extreme solvent stress. J Bacteriol 191:6773–6777
Sun S, Berg OG, Roth JR, Andersson DI (2009) Contribution of gene amplification to evolution of increased antibiotic resistance in Salmonella typhimurium. Genetics 182:1183–1195
Treves DS, Manning S, Adams J (1998) Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Mol Biol Evol 15:789–797
Wang L, Spira B, Zhou Z, Feng L, Maharjan RP, Li X, Li F, McKenzie C, Reeves PR, Ferenci T (2010) Divergence involving global regulatory gene mutations in an Escherichia coli population evolving under phosphate limitation. Genome Biol Evol 2:478–487
Warren RM, Victor TC, Streicher EM, Richardson M, van der Spuy GD, Johnson R, Chihota VN, Locht C, Supply P, van Helden PD (2004) Clonal expansion of a globally disseminated lineage of Mycobacterium tuberculosis with low IS6110 copy numbers. J Clin Microbiol 42:5774–5782
Whiteway J, Koziarz P, Veall J, Sandhu N, Kumar P, Hoecher B, Lambert IB (1998) Oxygen-insensitive nitroreductases: analyses of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J Bacteriol 180:5529–5539
Zhang Z, Saier MH Jr (2009) A novel mechanism of transposon-mediated gene activation. PLoS Genet 5(10):e1000689
Zhang Z, Yen MR, Saier MH Jr (2010) Precise excision of IS5 from the intergenic region between the fucPIK and the fucAO operons and mutational control of fucPIK operon expression in Escherichia coli. J Bacteriol 192:2013–2019
Zhou F, Olman V, Xu Y (2008) Insertion sequences show diverse recent activities in cyanobacteria and archea. BMC Genomics 9:36
Zinser ER, Schneider D, Blot M, Kolter R (2003) Bacterial evolution through the selective loss of beneficial genes: trade-offs in expression involving two loci. Genetics 164:1271–1277
Acknowledgments
We thank Thu Betteridge for technical support. This work was supported by the French Centre National de la Recherche Scientifique (CNRS), the University Joseph Fourier Grenoble, and the grant ANR-08-BLAN-0283-01 from the french Agence Nationale de la Recherche (ANR) program Blanc to D.S.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gaffé, J., McKenzie, C., Maharjan, R.P. et al. Insertion Sequence-Driven Evolution of Escherichia coli in Chemostats. J Mol Evol 72, 398–412 (2011). https://doi.org/10.1007/s00239-011-9439-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-011-9439-2