Abstract
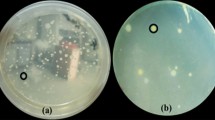
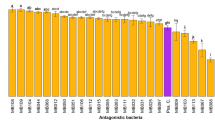
Sheath blight infection of rice by Rhizoctonia solani Kühn AG1-IA often results in serious yield losses in intensive rice cultivation. Biological control agents (BCAs) have previously been isolated but poor efficiency is often observed when applied under field conditions. This study compares a traditional dual-culture plate assay and a new water-surface microcosm assay for isolation of antagonistic soil bacteria. In the water-surface microcosm assay, floating pathogen mycelium is used as a source for isolation of hyphae-colonizing soil bacteria (HCSB), which are subsequently screened for antagonism. Ten antagonistic soil bacteria (ASB) isolated from a variety of Vietnamese rice soils using dual-culture plates were found to be affiliated with Bacillus based on 16S rRNA gene sequencing. However, all the ASB isolates grew poorly and showed no antagonism in the water-surface microcosm assay. In contrast, 11 (out of 13) HCSB isolates affiliated with Burkholderia sp. all grew well by colonizing the hyphae in the microcosms. Two of the Burkholderia sp. isolates, assigned to B. vietnamiensis based on recA gene sequencing, strongly inhibited fungal growth in both the dual-culture and water-surface microcosm assays; HCSB isolates affiliated to other species or species groups showed limited or no inhibition of R. solani in the microcosms. Our results suggest that HCSB obtained from floating pathogen hyphae can be a new source for isolation of efficient BCAs against R. solani, as the isolation assay mimics the natural habitat for fungal-bacterial interaction in the fields.



Similar content being viewed by others
References
Hashiba T, Kobayashi T (1996) Rice diseases incited by Rhizoctonia species. In: Sneh B, Jabaji-Hare S, Neate S, Dijst G (eds) Rhizoctonia species: taxomony, molecular biology, ecology, pathology and disease control. Kluwer Academic Publishers, Dordrecht, pp 331–340
Mew TW, Cottyn B, Pamplona R, Barrios H, Xiangmin L, Zhiyi C, Fan L, Nilpanit N, Arunyanart P, Kim PV, Du PV (2004) Applying rice seed-associated antagonistic bacteria to manage rice sheath blight in developing countries. Plant Dis 88:557–564
Mew TW, Leung H, Savary S, Cruz CMV, Leach JE (2004) Looking ahead in rice disease research and management. Crit Rev Plant Sci 23:103–127
Savary S, Castilla NP, Elazegui FA, McLaren CG, Ynalvez MA, Teng PS (1995) Direct and indirect effects of nitrogen supply and disease source structure on rice sheath blight spread. Phytopathology 85:959–965
Kobayashi T, Mew TW, Hashiba T (1997) Relationship between incidence of rice sheath blight and primary inoculum in the Philippines: mycelia in plant debris and sclerotia. Ann Phytopathol Soc Jpn 63:324–327
Lee FN, Rush MC (1983) Rice sheath blight: a major rice disease. Plant Dis 67:829–832
Willocquet L, Fernandez L, Savary S (2000) Effect of various crop establishment methods practised by Asian farmers on epidemics of rice sheath blight caused by Rhizoctonia solani. Plant Pathol 49:346–354
Gerhardson B (2002) Biological substitutes for pesticides. Trends Biotechnol 20:338–343
Whipps JM (2001) Microbial interactions and biocontrol in the rhizosphere. J Exp Bot 52:487–511
Compant S, Duffy B, Nowak J, Clément C, Barka EA (2005) Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. AEM 71:4951–4959
Kloepper JW (1996) Host specificity in microbe-microbe interactions. Bioscience 46:406–409
Kanjanamaneesathian M, Kusonwiriyawong C, Pengnoo A, Nilratana L (1998) Screening of potential bacterial antagonists for control of sheath blight in rice and development of suitable bacterial formulations for effective application. Australas Plant Pathol 27:198–206
Kazempour MN (2004) Biological control of Rhizoctoni solani, the causal agent of rice sheath blight by antagonistics bacteria in greenhouse and field conditions. Plant Pathol J 3:88–96
Luo JY, Xie GL, Li B, Luo YC, Zhao LH, Wang X, Liu B, Li W (2005) Gram-positive bacteria associated with rice in China and their antagonists against the pathogens of sheath blight and bakanae disease in rice. Rice Sci 12:213–218
Haas D, Défago G (2005) Biological control of soil-borne pathogens by fluorescent Pseudomonads. Nat Rev Microbiol 3:307–319
Alabouvette C, Olivain C, Steinberg C (2006) Biological control of plant diseases: the European situation. Eur J Plant Pathol 114:329–341
Quan CS, Zheng W, Liu Q, Ohta Y, Fan SD (2006) Isolation and characterization of a novel Burkholderia cepacia with strong antifungal activity against Rhizoctonia solani. Appl Microbiol Biotechnol 72:1276–1284
Whipps JM (1987) Effect of media on growth and interactions between a range of soil borne glasshouse pathogens and antagonistic fungi. New Phytol 107:127–142
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA (1990) Combination of 16S rRNA-targeted oligonucleotide probes with flow-cytometry for analyzing mixed microbial-populations. Appl Environ Microbiol 56:1919–1925
Lane DJ (1991) 16 S/23 S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, pp 115–175
Mahenthiralingam E, Bischof J, Byrne SK, Radomski C, Davies JE, Av-Gay Y, Vandamme P (2000) DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J Clin Microbiol 38:3165–3173
de Souza JT, Raaijmakers JM (2003) Polymorphisms within the prnD and pltC genes from pyrrolnitrin and pyoluteorin-producing Pseudomonas and Burkholderia spp. FEMS Microbiol Ecol 43:21–34
Svercel M, Duffy B, Défago G (2007) PCR amplification of hydrogen cyanide biosynthetic locus hcnAB in Pseudomonas spp. J Microbiol Meth 70:209–213
Hoa NM, Singh U, Samonte HP (1998) Potassium supplying capacity of some lowland rice soils in the Mekong delta. Better Crops Int 12:11–15
Karlin S, Weinstock GM, Brendel V (1995) Bacterial classifications derived from RecA protein-sequence comparisons. J Bacteriol 177:6881–6893
Eisen JA (1995) The RecA protein as a model molecule for molecular systematic studies of bacteria: comparison of trees of RecAs and 16 S rRNAs from the same species. J Mol Evol 41:1105–1123
Kim HS, Park J, Choi SW, Choi KH, Lee GP, Ban SJ, Lee CH, Kim CS (2003) Isolation and characterization of Bacillus strains for biological control. J Microbiol 41:196–201
Cazorla FM, Romero D, Pérez-García A, Lugtenberg BJJ, de Vicente A, Bloemberg G (2007) Isolation and characterization of antagonistic Bacillus subtilis strains from the avocado rhizoplane displaying biocontrol activity. J Appl Microbiol 103:1950–1959
Hu QP, Xu JG, Song P, Song JN, Chen WL (2008) Isolation and identification of a potential biocontrol agent Bacillus subtilis QM3 from Qinghai yak dung in China. World J Microbiol Biotechnol 24:2451–2458
Hervas A, Casamayor EO (2009) High similarity between bacterioneuston and airborne bacterial community compositions in a high mountain lake area. FEMS Microbiol Ecol 67:219–228
Timonen S, Jørgensen KS, Haahtela K, Sen R (1998) Bacterial community structure at defined locations of Pinus sylvestris - Suillus bovinus and Pinus sylvestris—Paxillus involutus mycorrhizospheres in dry pine forest humus and nursery peat. Can J Microbiol 44:499–513
Mansfeld-Giese K, Larsen J, Bødker L (2002) Bacterial populations associated with mycelium of the arbuscular mycorrhizal fungus Glomus intraradices. FEMS Microbiol Ecol 41:133–140
Aspray TJ, Jones EE, Whipps JM, Bending GD (2006) Importance of mycorrhization helper bacteria cell density and metabolite localization for the Pinus sylvestris-Lactarius rufus symbiosis. FEMS Microbiol Ecol 56:25–33
Toljander JF, Artursson V, Paul LR, Jansson JK, Finlay RD (2006) Attachment of different soil bacteria to arbuscular mycorrhizal fungal extraradical hyphae is determined by hyphal vitality and fungal species. FEMS Microbiol Lett 254:34–40
Scheublin TR, Sanders IR, Keel C, van der Meer JR (2010) Characterisation of microbial communities colonising the hyphal surfaces of arbuscular mycorrhizal fungi. ISME J 4:752–763
Jha B, Thakur MC, Gontia I, Albrecht V, Stoffels M, Schmid M, Hartmann A (2009) Isolation, partial identification and application of diazotrophic rhizobacteria from traditional Indian rice cultivars. Eur J Soil Biol 45:62–72
Kalwaslińska A, Kesy J, Donderski W (2008) Biodegradation of carbendazim by epiphytic and neustonic bacteria of eutrophic Chełmzyńskie. Pol J Microbiol 57:221–230
Cartwright DK, Chilton WS, Benson DM (1995) Pyrrolnitrin and phenazine production by Pseudomonas cepacia, strain 5.5B, a biocontrol agent of Rhizoctonia solani. Appl Microbiol Biotechnol 43:211–216
Rosales AM, Thomashow L, Cook RJ, Mew TW (1995) Isolation and identification of antifungal metabolites produced by rice-associated antagonistic Pseudomonas spp. Phytopathology 85:1028–1032
EL-Banna N, Winkelmann G (1998) Pyrrolnitrin from Burkholderia cepacia: antibiotic activity against fungi and novel activities against streptomycetes. J Appl Microbiol 85:69–78
Jiao Y, Yoshihara T, Ishikuri S, Uchino H, Ichihara A (1996) Structural identification of cepaciamide A, a novel fungitoxic compound from Pseudomonas cepacia D-202. Tetrahedron Lett 37:1039–1042
Mao S, Lee SJ, Hwangbo H, Kim YW, Park KH, Cha GS, Park RD, Kim KY (2006) Isolation and characterization of antifungal substances from Burkholderia sp. culture broth. Curr Microbiol 53:358–364
Nielsen MN, Sørensen J (1999) Chitinolytic ativity of Pseudomonas fluorescens isolates from barley and sugar beet rhizosphere. FEMS Microbiol Ecol 30:217–227
Ogawa K, Yoshida N, Kariya K, Ohnishi C, Ikeda R (2002) Purification and characterization of a novel chitinase from Burkholderia cepacia strain KH2 isolated from the bed log of Lentinus edodes, Shiitake mushroom. J Gen Appl Microbiol 48:25–33
Payne GW, Vandamme P, Morgan SH, LiPuma JJ, Coenye T, Weightman AJ, Jones TH, Mahenthiralingam E (2005) Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl Environ Microbiol 71:3917–3927
Gillis M, Van TV, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez MP (1995) Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol 45:274–289
Schmidt S, Blom JF, Pernthaler J, Berg G, Baldwin A, Mahenthiralingam E, Eberl L (2009) Production of the antifungal compound pyrrolnitrin is quorum sensing-regulated in members of the Burkholderia cepacia complex. Environ Microbiol 11:1422–1437
Chiarini L, Bevivino A, Dalmastri C, Tabacchioni S, Visca P (2006) Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol 14:277–286
Vanlaere E, LiPuma JJ, Baldwin A, Henry D, De Brandt E, Mahenthiralingam E, Speert D, Dowson C, Vandamme P (2008) Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int J Syst Evol Microbiol 58:1580–1590
Anandham R, Indira Gandhi P, Kwon SW, Sa TM, Kim YK, Jee HJ (2009) Mixotrophic metabolism in Burkholderia kururiensis subsp. thiooxydans subsp. nov., a facultative chemolithoautotrophic thiosulfate oxidizing bacterium isolated from rhizosphere soil and proposal for classification of the type strain of Burkholderia kururiensis as Burkholderia kururiensis subsp. kururiensis subsp. nov. Arch Microbiol 191:885–894
Yang JH, Liu HX, Zhu GM, Pan YL, Xu LP, Guo JH (2008) Diversity analysis of antagonists from rice-associated bacteria and their application in biocontrol of rice diseases. J Appl Microbiol 104:91–104
Cottyn B, Debode J, Regalado E, Mew TW, Swings J (2009) Phenotypic and genetic diversity of rice seed-associated bacteria and their role in pathogenicity and biological control. J Appl Microbiol 107:885–897
Jeger MJ, Jeffries P, Elad Y, Xu XM (2009) A generic theoretical model for biological control of foliar plant diseases. J Theor Biol 256:201–214
Acknowledgment
This study was supported by a research grant from the Danish Ministry of Foreign Affairs on “Integrated disease and nutrient management in intensive rice production systems in Vietnam” (104.DAN.8.L.727). We thank Dorthe Ganzhorn and Kirsten Henriksen for excellent laboratory assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cuong, N.D., Nicolaisen, M.H., Sørensen, J. et al. Hyphae-Colonizing Burkholderia sp.—A New Source of Biological Control Agents Against Sheath Blight Disease (Rhizoctonia solani AG1-IA) in Rice. Microb Ecol 62, 425–434 (2011). https://doi.org/10.1007/s00248-011-9823-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-011-9823-x