Abstract
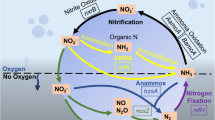
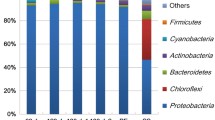
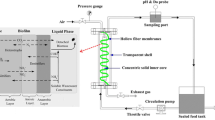
Nitrate is a serious problem in closed-circuit public aquariums because its accumulation rapidly becomes toxic to many lifeforms. A moving bed biofilm denitrification reactor was installed at the Montreal Biodome to treat its 3,250-m3 seawater system. Naturally occurring microorganisms from the seawater affluent colonized the reactor carriers to form a denitrifying biofilm. Here, we investigated the functional diversity of this biofilm by retrieving gene sequences related to narG, napA, nirK, nirS, cnorB, and nosZ. A total of 25 sequences related to these genes were retrieved from the biofilm. Among them, the corresponding napA1, nirK1, cnorB9, and nosZ3 sequences were identical to the corresponding genes found in Hyphomicrobium sp. NL23 while the narG1 and narG2 sequences were identical to the two corresponding narG genes found in Methylophaga sp. JAM1. These two bacterial strains were previously isolated from the denitrifying biofilm. To assess the abundance of denitrifiers and nitrate respirers in the biofilm, the gene copy number of all the narG, napA, nirS, and nirK sequences found in biofilm was determined by quantitative PCR. napA1, nirK1, narG1, and narG2, which were all associated with either Methylophaga sp. JAM1 or Hyphomicrobium sp. NL23, were the most abundant genes. The other genes were 10 to 10,000 times less abundant. nirK, cnorB, and nosZ but not napA transcripts from Hyphomicrobium sp. NL23 were detected in the biofilm, and only the narG1 transcripts from Methylophaga sp. JAM1 were detected in the biofilm. Among the 19 other genes, the transcripts of only two genes were detected in the biofilm. Our results show the predominance of Methylophaga sp. JAM1 and Hyphomicrobium sp. NL23 among the denitrifiers detected in the biofilm. The results suggest that Hyphomicrobium sp. NL23 could use the nitrite present in the biofilm generated by nitrate respirers such as Methylophaga sp. JAM1.

Similar content being viewed by others
References
Auclair J, Lépine F, Parent S, Villemur R (2010) Dissimilatory reduction of nitrate in seawater by a Methylophaga strain containing two highly divergent narG sequences. ISME J 4:1302–1313
Balderston WL, Sieburth JM (1976) Nitrate removal in closed-system aquaculture by columnar denitrification. Appl Environ Microbiol 32:808–818
Blasco F, Iobbi C, Ratouchniak J, Bonnefoy V, Chippaux M (1990) Nitrate reductases of Escherichia coli: sequence of the second nitrate reductase and comparison with that encoded by the narGHJI operon. Mol Gen Genet 222:104–111
Braker G, Fesefeldt A, Witzel KP (1998) Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol 64:3769–3775
Braker G, Tiedje JM (2003) Nitric oxide reductase (norB) genes from pure cultures and environmental samples. Appl Environ Microbiol 69:3476–3483
Bru D, Sarr A, Philippot L (2007) Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments. Appl Environ Microbiol 73:5971–5974
Camargo JA, Alonso A, Salamanca A (2005) Nitrate toxicity to aquatic animals: a review with new data for freshwater invertebrates. Chemosphere 58:1255–1267
Chang L, Wei LLC, Audia JP, Morton RA, Schellhorn HE (1999) Expression of the Escherichia coli NRZ nitrate reductase is highly growth phase dependent and is controlled by RpoS, the alternative vegetative sigma factor. Mol Microbiol 34:756–766
Cortes-Lorenzo C, Molina-Munoz ML, Gomez-Villalba B, Vilchez R, Ramos A, Rodelas B, Hontoria E, Gonzalez-Lopez J (2006) Analysis of community composition of biofilms in a submerged filter system for the removal of ammonia and phenol from industrial wastewater. Biochem Soc Trans 34:165–168
Drysdale GD, Kasan HC, Bux F (2001) Assessment of denitrification by the ordinary heterotrophic organisms in an NDBEPR activated sludge sytem. Water Sci Technol 43:147–154
Enticknap JJ, Kelly M, Peraud O, Hill RT (2006) Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl Environ Microbiol 72:3724–3732
Felsenstein J (1989) PHYLIP—phylogeny inference package. Cladistics 5:164–166
Fesefeldt A, Kloos K, Bothe H, Lemmer H, Gliesche CG (1998) Distribution of denitrification and nitrogen fixation genes in Hyphomicrobium spp. and other budding bacteria. Can J Microbiol 44:181–186
Flanagan DA, Gregory LG, Carter JP, Karakas-Sen A, Richardson DJ, Spiro S (1999) Detection of genes for periplasmic nitrate reductase in nitrate respiring bacteria and in community DNA. FEMS Microbiol Lett 177:263–270
Gentile ME, Jessup CM, Nyman JL, Criddle CS (2007) Correlation of functional instability and community dynamics in denitrifying dispersed-growth reactors. Appl Environ Microbiol 73:680–690
Ginige MP, Hugenholtz P, Daims H, Wagner M, Keller J, Blackall LL (2004) Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization-microautoradiography to study a methanol-fed denitrifying microbial community. Appl Environ Microbiol 70:588–596
Gregory LG, Karakas-Sen A, Richardson DJ, Spiro S (2000) Detection of genes for membrane-bound nitrate reductase in nitrate-respiring bacteria and in community DNA. FEMS Microbiol Lett 183:275–279
Grguric G, Wetmore SS, Fournier RW (2000) Biological denitrification in a closed seawater system. Chemosphere 40:549–555
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hallin S, Lindgren PE (1999) PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl Environ Microbiol 65:1652–1657
Hallin S, Throbäck IN, Dicksved J, Pell M (2006) Metabolic profiles and genetic diversity of denitrifying communities in activated sludge after addition of methanol or ethanol. Appl Environ Microbiol 72:5445–5452
Henry S, Baudoin E, Lopéz-Gutiérrez JC, Martin-Laurent F, Brauman A, Philippot L (2004) Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J Microbiol Methods 59:327–335
Jones CM, Hallin S (2010) Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. ISME J 4:633–641
Kandeler E, Deiglmayr K, Tscherko D, Bru D, Philippot L (2006) Abundance of narG, nirS, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl Environ Microbiol 72:5957–5962
Kloos K, Fesefeldt A, Gliesche CG, Bothe H (1995) DNA-probing indicates the occurrence of denitrification and nitrogen fixation genes in Hyphomicrobium. Distribution of denitrifying and nitrogen fixing isolates of Hyphomicrobium in a sewage treatment plant. FEMS Microb Ecol 18:205–213
Labbé N, Juteau P, Parent S, Villemur R (2003) Bacterial diversity in a marine methanol-fed denitrification reactor at the Montreal Biodome, Canada. Microb Ecol 46:12–21
Labbé N, Laurin V, Juteau P, Parent S, Villemur R (2007) Microbiological community structure of the biofilm of a methanol-fed, marine denitrification system, and identification of the methanol-utilizing microorganisms. Microb Ecol 53:621–630
Labbé N, Parent S, Villemur R (2003) Addition of trace metals increases denitrification rate in closed marine systems. Water Res 37:914–920
Lange R, Hengge-Aronis R (1991) Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol 5:49–59
Laurin V, Labbé N, Juteau P, Parent S, Villemur R (2006) Long-term storage conditions for carriers with denitrifying biomass of the fluidized, methanol-fed denitrification reactor of the Montreal Biodome, and the impact on denitrifying activity and bacterial population. Water Res 40:1836–1840
Levy-Booth DJ, Winder RS (2010) Quantification of nitrogen reductase and nitrite reductase genes in soil of thinned and clear-cut Douglas-fir stands by using real-time PCR. Appl Environ Microbiol 76:7116–7125
Moran MA, Belas R, Schell MA, González JM, Sun F, Sun S, Binder BJ, Edmonds J, Ye W, Orcutt B, Howard EC, Meile C, Palefsky W, Goesmann A, Ren Q, Paulsen I, Ulrich LE, Thompson LS, Saunders E, Buchan A (2007) Ecological genomics of marine roseobacters. Appl Environ Microbiol 73:4559–4569
Neef A, Zaglauer A, Meier H, Amann R, Lemmer H, Schleifer KH (1996) Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl Environ Microbiol 62:4329–4339
Neufeld JD, Driscoll BT, Knowles R, Archibald FS (2001) Quantifying functional gene populations: comparing gene abundance and corresponding enzymatic activity using denitrification and nitrogen fixation in pulp and paper mill effluent treatment systems. Can J Microbiol 47:925–934
Palmer K, Drake HL, Horn MA (2009) Genome-derived criteria for assigning environmental narG and nosZ sequences to operational taxonomic units of nitrate reducers. Appl Environ Microbiol 75:5170–5174
Parent S, Morin A (2000) N budget as water quality management tool in closed aquatic mesocosms. Water Res 34:1846–1856
Richardson DJ, Berks BC, Russell DA, Spiro S, Taylor CJ (2001) Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell Mol Life Sci 58:165–178
Sauthier N, Grasmick A, Blancheton JP (1998) Biological denitrification applied to a marine closed aquaculture system. Water Res 32:1932–1938
Scala DJ, Kerkhof LJ (1998) Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol Lett 162:61–68
Scala DJ, Kerkhof LJ (1999) Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl Environ Microbiol 65:1681–1687
Serghini MA, Ritzenthaler C, Pinck L (1989) A rapid and efficient miniprep for isolation of plasmid DNA. Nucleic Acids Res 17:3604–3604
Shapleigh J (2006) The denitrifying prokaryotes. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The prokaryotes. Springer, New York, pp 769–792
Sperl GT, Hoare DS (1971) Denitrification with methanol: a selective enrichment for Hyphomicrobium species. J Bacteriol 108:733–736
Tal Y, Nussinovitch A, van Rijn J (2003) Nitrate removal in aquariums by immobilized Pseudomonas. Biotechnol Prog 19:1019–1021
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Visvanathan C, Phong DD, Jegatheesan V (2008) Hydrogenotrophic denitrification of highly saline aquaculture wastewater using hollow fiber membrane bioreactor. Environ Technol 29:701–707
Yoon JH, Oh TK, Park YH (2004) Kangiella koreensis gen. nov., sp. nov. and Kangiella aquimarina sp. nov., isolated from a tidal flat of the Yellow Sea in Korea. Int J Syst Evol Microbiol 54:1829–1835
Yoshie S, Makino H, Hirosawa H, Shirotani K, Tsuneda S, Hirata A (2006) Molecular analysis of halophilic bacterial community for high-rate denitrification of saline industrial wastewater. Appl Microbiol Biotechnol 72:182–189
Yoshie S, Noda N, Miyano T, Tsuneda S, Hirata A, Inamori Y (2001) Microbial community analysis in the denitrification process of saline-wastewater by denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA and the cultivation method. J Biosci Bioeng 92:346–353
Yoshie S, Noda N, Tsuneda S, Hirata A, Inamori Y (2004) Salinity decreases nitrite reductase gene diversity in denitrifying bacteria of wastewater treatment systems. Appl Environ Microbiol 70:3152–3157
Zumft WG (1997) Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61:533–616
Acknowledgments
This research was supported by the Montreal Biodome and a grant to R.V. from the Natural Sciences and Engineering Research Council of Canada. J.A. held scholarships from the Fondation Armand-Frappier and the Fonds de recherche sur la nature et les technologies du Québec. We are grateful to Maxime Hémond and the engineering staff of the Biodome for their technical support. This manuscript was revised by Elsevier Language Editing Services.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Auclair, J., Parent, S. & Villemur, R. Functional Diversity in the Denitrifying Biofilm of the Methanol-Fed Marine Denitrification System at the Montreal Biodome. Microb Ecol 63, 726–735 (2012). https://doi.org/10.1007/s00248-011-9960-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-011-9960-2