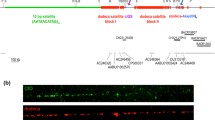
Abstract
The transcriptional framework of the eukaryotic centromere core has been described in budding yeast and rice, but for most eukaryotes and all vertebrates it remains largely unknown. The lack of large pericentric repeats in the tammar wallaby has made it possible to map and identify the transcriptional units at the centromere in a mammalian species for the first time. We show that these transcriptional units, comprised of satellites and a retrovirus, are bound by centromere proteins and that they are the source of a novel class of small RNA. The endogenous retrovirus from which these small RNAs are derived is now known to be in the centromere domain of several vertebrate classes. The discovery of this new RNA form brings together several independent lines of evidence that point to a conserved retroviral-encoded processed RNA entity within eukaryotic centromeres.






Similar content being viewed by others
References
Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ (2007) Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316:744–747
Blower MD, Sullivan BA, Karpen GH (2002) Conserved organization of centromeric chromatin in flies and humans. Developmental Cell 2:319–330
Bouzinba-Segard H, Guais A, Francastel C (2006) Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci U S A 103:8709–8714
Boyd KE, Farnham PJ (1999) Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol Cell Biol 19:8393–8399
Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128:1089–1103
Bulazel K, Metcalfe C, Ferreri GC, Yu J, Eldridge MD, O’Neill RJ (2006) Cytogenetic and molecular evaluation of centromere-associated DNA sequences from a marsupial (Macropodidae: Macropus rufogriseus) X chromosome. Genetics 172:1129–1137
Bulazel K, Ferreri GC, Eldridge MD, O’Neill RJ (2007) Species-specific shifts in centromere sequence composition are coincident with breakpoint reuse in karyotypically divergent lineages. Genome Biol 8:R170
Cheng Z, Dong F, Langdon T, Ouyang S, Buell CR, Gu M, Blattner FR, Jiang J (2002) Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14:1691–1704
Choo K (1997) The centromere. Oxford University Press, Oxford
Dalby B, Cates S, Harris A, Ohki EC, Tilkins ML, Price PJ, Ciccarone VC (2004) Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods 33:95–103
Diaz MO, Barsacchi-Pilone G, Mahon KA, Gall JG (1981) Transcripts from both strands of a satellite DNA occur on lampbrush chromosome loops of the newt Notophthalmus. Cell 24:649–659
Dunn CA, Romanish MT, Gutierrez LE, van de Lagemaat LN, Mager DL (2006) Transcription of two human genes from a bidirectional endogenous retrovirus promoter. Gene 366:335–342
Earnshaw WC, Rothfield N (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91:313–321
Ferreri GC, Marzelli M, Rens W, O'Neill RJ (2004) A centromere-specific retroviral element associated with breaks of synteny in macropodine marsupials. Cytogenet Genome Res 107:115–118
Ferreri GC, Liscinsky DM, Mack JA, Eldridge MD, O’Neill RJ (2005) Retention of latent centromeres in the Mammalian genome. J Hered 96:217–224
Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M (2004) Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol 6:784–791
Hammond SM, Bernstein E, Beach D, Hannon GJ (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293–296
Hartig JV, Tomari Y, Forstemann K (2007) piRNAs—the ancient hunters of genome invaders. Genes Dev 21:1707–1713
Hayman DL (1977) Chromosome number-constancy and variation. In: Gilmore D (ed) The biology of marsupials. Macmillan, London
Hayman DL (1990) Marsupial cytogenetics. In: Cooper DW (ed) Mammals from pouches and eggs: genetics, breeding and evolution of marsupials and monotremes. CSIRO, Melbourne
Hudson DF, Fowler KJ, Earle E, Saffery R, Kalitsis P, Trowell H, Hill J, Wreford NG, de Kretser DM, Cancilla MR, Howman E, Hii L, Cutts SM, Irvine DV, Choo KH (1998) Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J Cell Biol 141:309–319
Jiang J, Birchler JA, Parrott WA, Dawe RK (2003) A molecular view of plant centromeres. Trends Plant Sci 8:570–575
Josse T, Teysset L, Todeschini A-L, Sidor CM, Anxolabehere D, Ronsseray S (2007) Telomeric trans-silencing: an epigenetic repression combining RNA silencing and heterochromatin formation. PLOS Genet 3:e158
Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K (2005) Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19:489–501
Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316:1484–1488
Kim VN (2006) Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev 20:1993–1997
Kuznetsova I, Podgornaya O, Ferguson-Smith MA (2006) High-resolution organization of mouse centromeric and pericentromeric DNA. Cytogenet Genome Res 112:248–255
Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE (2006) Characterization of the piRNA complex from rat testes. Science 313:363–367
Lee Y, Jeon K, Lee JT, Kim S, Kim VN (2002) MicroRNA maturation: stepwise processing and subcellular localization. Embo J 21:4663–4670
Lee HR, Neumann P, Macas J, Jiang J (2006) Transcription and evolutionary dynamics of the centromeric satellite repeat CentO in rice. Mol Biol Evol 23:2505–2520
Lichter P, Cremer T, Borden J, Manuelidis L, Ward DC (1988) Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Human Genet 80:224–234
May BP, Lippman ZB, Fang Y, Spector DL, Martienssen RA (2005) Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet 1:e79
Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ (2005) Characterization of dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A 102:12135–12140
Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36:138–145
Neumann P, Yan H, Jiang J (2007) The centromeric retrotransposons of rice are transcribed and differentially processed by RNA interference. Genetics 176:749–761
Ohzeki J, Nakano M, Okada T, Masumoto H (2002) CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol 159:765–775
Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H (2007) CENP-B controls centromere formation depending on the chromatin context. Cell 131:1287–1300
O’Neill RJ, O'Neill MJ, Graves JA (1998) Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393:68–72
Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC (2004) Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303:669–672
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Reinhart BJ, Bartel DP (2002) Small RNAs correspond to centromere heterochromatic repeats. Science 297:1831
Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD (2003) Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell 12:1591–1598
Sambrook J, Fritsch EF, Maniatis T (2001) Molecular cloning. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF (2001) Genomic and genetic definition of a functional human centromere. Science 294:109–115
Topp CN, Zhong CX, Dawe RK (2004) Centromere-encoded RNAs are integral components of the maize kinetochore. Proc Natl Acad Sci U S A 101:15986–15991
Ugarkovic D (2005) Functional elements residing within satellite DNAs. EMBO Rep 6:1035–1039
Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD (2006) A distinct small RNA pathway silences selfish genetic elements in the germ line. Science 313:320–324
Valgardsdottir R, Chiodi I, Giordano M, Cobianchi F, Riva S, Biamonti G (2005) Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol Biol Cell 16:2597–2604
Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303:672–676
Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833–1837
Volpe T, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, Allshire RC (2003) RNA interference is required for normal centromere function in fission yeast. Chromosome Res 11:137–146
White SA, Allshire RC (2004) Loss of dicer fowls up centromeres. Nat Cell Biol 6:696–697
Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, Choo KH (2007) Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res 17:1146–1160
Acknowledgements
We thank Judy Brown and Andrew Pask for critical evaluation of this manuscript and Marilyn Renfree and Andrew Pask for tammar wallaby material. This work was supported by a grant from the UCONN Research Foundation and an National Science Foundation award to RO.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: R. Allshire
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Calibration for fiber FISH. a M. musculus chromosome 11 BAC (RP23-234K24, accession number AL596181) mapped to fibers (blue gray) of M. musculus; b and c M. eugenii chromosome 6 BAC (ME_Kba-583J6, accession number AC166217) mapped to fibers (blue gray) of M. eugenii. Both probes were detected with avidin–FITC (green) (GIF 98 KB)
Fig. S2
CREST and CENP-B recognize centromeres of M. eugenii. A partial cell, containing both compact centromeric chromatin and extended centromere chromatin fibers (panel 1, DAPI-stained, blue gray) detected with CREST anticentromere antibodies (panel 2, antihuman Texas Red) and anti-CENP-B (panel 3, antimouse FITC, green). While both antibodies detected compact chromatin (bottom left of panels 2–4), only CREST recognized extended chromatin fibers (indicated with an arrow). The merge of all three wavelengths is shown in panel 4 (GIF 38.3 KB)
Fig. S3
The CENP-B binding domain of the unbound ChIP product is highly mutated. The unbound ChIP sat23 product obtained from chromatin immunoprecipitation with an antibody to CENP-B contains three 1-bp mismatches (highlighted in bold) compared to the tammar wallaby CENP-B box consensus (top) and sat23 sequence obtained from the bound fraction (CenpB bound sat23; bottom). The bases critical to binding of CENP-B are highlighted in red (Bulazel et al. 2006). The presence of mismatches within this conserved binding domain indicates that the sequence obtained in the unbound fraction has lost its ability to bind CENP-B (Bulazel et al. 2006) and is most likely located more distal to the centromere core (GIF 16.9 KB)
Fig. S4
Small RNA are homologous to KERV-1 LTR and KERV-1 gag and sat23. Size-fractionated small RNA pool was dephosphorylated, end-labeled with γ32P, and hybridized to Southern blots containing PCR products corresponding to KERV-1 LTR (lane 1), sat23 (lane 2), and KERV-1 gag (lane 3) indicating that elements residing at the centromere produce small RNA. A noncentromeric sequence (B15) for the tammar wallaby (Bulazel et al. 2007) was included as a negative control (lane 4) (GIF 23.8 KB)
Rights and permissions
About this article
Cite this article
Carone, D.M., Longo, M.S., Ferreri, G.C. et al. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma 118, 113–125 (2009). https://doi.org/10.1007/s00412-008-0181-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-008-0181-5