Abstract
The DNA-dependent adenosine triphosphatase (ATPase) Plk1-interacting checkpoint helicase (PICH) has recently been implicated in spindle checkpoint (SAC) signaling (Baumann et al., Cell 128(1):101–114, 2007). Depletion of PICH by siRNA abolished the SAC and resulted in an apparently selective loss of Mad2 from kinetochores, suggesting a role for PICH in the regulation of the Mad1–Mad2 interaction. An apparent rescue of SAC functionality by overexpression of PICH in PICH-depleted cells initially seemed to confirm a role for PICH in the SAC. However, we have subsequently discovered that all PICH-directed siRNA oligonucleotides that abolish the SAC also reduce Mad2 mRNA and protein expression. This reduction is functionally significant, as PICH siRNA does not abolish SAC activity in a cell line that harbors a bacterial artificial chromosome driving the expression of murine Mad2. Moreover, we identified several siRNA duplexes that effectively deplete PICH but do not significantly affect SAC functionality or Mad2 abundance or localization. Finally, we discovered that the ability of overexpressed PICH to restore SAC activity in PICH-depleted cells depends on sequestration of the mitotic kinase Plk1 rather than ATPase activity of PICH, pointing to an underlying mechanism of “bypass suppression.” In support of this view, depletion or inhibition of Plk1 also rescued SAC activity in cells harboring low levels of Mad2. This observation suggests that a reduction of Plk1 activity partially compensates for reduced Mad2 levels and argues that Plk1 normally reduces the strength of SAC signaling. Collectively, our results question the role of PICH in the SAC and instead identify Mad2 as a sensitive off target for small RNA duplexes. In support of the latter conclusion, our evidence suggests that an off-target effect on Mad2 may also contribute to explain the apparent role of the Tao1 kinase in SAC signaling (Draviam et al., Nat Cell Biol 9(5):556–564, 2007).
Similar content being viewed by others
Introduction
The DNA-dependent adenosine triphosphatase (ATPase) Plk1-interacting checkpoint helicase (PICH) was discovered as a binding partner and substrate of the mitotic kinase Plk1 (Baumann et al. 2007). During early mitosis, PICH is concentrated at the centromere/kinetochore (KT) region of mitotic chromosomes. In response to inhibition or depletion of Plk1, PICH spreads over chromosome arms, indicating that its localization is controlled by Plk1 activity (Baumann et al. 2007). Conversely, PICH apparently contributes to the recruitment of Plk1 to chromosome arms (Santamaria et al. 2007; Leng et al. 2008). Most interestingly, PICH associates with ultrafine DNA bridges (UFBs) that often connect the KTs of separating sister chromatids (Baumann et al. 2007; Wang et al. 2008). After inactivation of the SAC and cleavage of centromere-associated cohesin complexes by separase (Hauf et al. 2001; Uhlmann et al. 1999), PICH-positive UFBs elongate concomitant with sister chromatid separation and often reach lengths of several microns before they are finally resolved, presumably through the action of topoisomerase II (Baumann et al. 2007; Wang et al. 2008). While PICH presently constitutes the most universal marker for UFBs, it is intriguing that Bloom syndrome RecQ helicase as well as its complex partners RMI1 and topoisomerase III also associate with some PICH-positive structures (Chan et al. 2007; Bachrati and Hickson 2008). Moreover, a subset of noncentromeric UFBs was recently found to connect fragile site loci associated with Fanconi anemia proteins FANCD2 and FANCI, and the available evidence suggests that these latter UFBs reflect abnormal intertwined DNA structures induced by replicative stress (Chan et al. 2009; Naim and Rosselli 2009).
The conspicuous localization of PICH to the centromere/KT region originally suggested a role for this DNA-dependent ATPase in the SAC. In support of this idea, siRNA-mediated depletion of PICH by two different siRNA oligonucleotides (PICH-1 and PICH-2) abolished the checkpoint (Baumann et al. 2007), and results consistent with SAC failure were subsequently observed after depletion of PICH by a third siRNA oligonucleotide (hereafter referred to as PICH-CC; Leng et al. 2008). Furthermore, siRNA-mediated PICH depletion caused the apparently selective loss of the checkpoint protein Mad2 from KTs, while the localization of the Mad2-binding partner Mad1 was unaffected (Baumann et al. 2007). Independently, an apparently selective loss of Mad2 from KTs was reported in response to siRNA-mediated depletion of the protein kinase Tao1, whose apparent role in the SAC was discovered in a functional genomic screen (Draviam et al. 2007). At face value, these observations suggested that both PICH and Tao1 are required for SAC activity and that these two proteins might cooperate in regulating the Mad1–Mad2 interaction at KTs.
The present study was undertaken with the aim of elucidating the molecular mechanism underlying the purported checkpoint function of PICH. Although this function appeared to be supported by concordant results obtained with three different siRNA oligonucleotides, the studies described here lead us to conclude that the checkpoint failure formerly attributed to the depletion of PICH most likely reflects an off-target effect that causes the lowering of Mad2 transcript and protein. Our data further suggest that an off-target effect may similarly explain the purported checkpoint function of the Tao1 kinase. The direct (off) target of the various siRNA oligonucleotides examined here is unlikely to be the Mad2 transcript itself; instead, our results suggest that the regulatory network controlling Mad2 expression represents a particularly sensitive target for small RNAs. Interestingly, the off-target effect of a PICH-directed siRNA oligonucleotide could be rescued by overexpression of PICH, which a priori would seem to demonstrate specificity. However, we discovered that this apparent rescue required the ability of the overexpressed protein to sequester the mitotic kinase Plk1. A similar, albeit partial “rescue” could be accomplished by depletion or inhibition of Plk1, thus providing an example for the apparent “rescue” of a siRNA phenotype via bypass suppression.
Results and discussion
Re-examination of the proposed SAC function of PICH
PICH could effectively be depleted from HeLa cells using any one of three different siRNA oligonucleotides, referred to as PICH-1, PICH-2, and PICH-CC. Live-cell imaging performed on HeLa cells stably expressing histone H2B–green fluorescent protein (GFP) showed that all three siRNA treatments also caused remarkably similar mitotic phenotypes. Compared to control siRNA (siGL2)-treated cells, PICH-depleted cells failed to form proper metaphase plates and instead displayed premature onset of anaphase (Supplementary Fig. 1a); moreover, PICH-depleted cells failed to arrest in response to nocodazole (Supplementary Fig. 1b), confirming and extending previous results (Baumann et al. 2007). To strengthen the conclusion that PICH is the critical target of siRNA in these experiments, we asked whether normal mitotic timing could be restored in PICH-1 siRNA-treated cells by overexpression of an siRNA-resistant PICH protein tagged with mCherry. Whereas control cells spent an average time of 43.8 min between nuclear envelope breakdown (NEBD) and anaphase onset, PICH-depleted cells expressing mCherry vector alone expeditiously initiated anaphase with an average time of 26.8 min (Fig. 1). In contrast, the overexpression of mCherry-tagged PICH wild type (WT) successfully restored the mitotic timing to near-normal levels (42.6 min), although some cells died, in line with the previously observed toxicity of excess PICH (Fig. 1; Baumann et al. 2007). The concordant phenotypes produced by three independent siRNA oligonucleotides and the apparently successful rescue initially bolstered our confidence that PICH plays an essential role in SAC signaling. However, several observations subsequently challenged this conclusion, as described in the following sections.
Overexpression of PICH in PICH siRNA-treated cells restores mitotic timing. HeLa cells stably expressing histone H2B-GFP were transfected simultaneously with the indicated siRNA duplexes and plasmids and analyzed by time-lapse microscopy. mCherry-positive cells were analyzed to determine the average time from NEBD to anaphase onset. Left panels show single-plane images selected from movies, and time stamps indicate hours:minutes. Right panels show representative fate maps of 25 individual cells (the whole experiment being repeated independently three times), with red columns indicating the time span from NEBD to anaphase onset and blue columns indicating cells that died in interphase during imaging. Average times from NEBD to anaphase onset are indicated to the right
A first unexpected result emerged when we generated a 293T cell line (293T TREX-shPICH), which allowed the inducible expression of a short hairpin RNA (shRNA) coding for the PICH-1 siRNA sequence. Three stable clones were obtained, all of which showed efficient PICH depletion after 72 h of incubation with tetracycline. This depletion was reversible, as PICH protein was partially restored upon removal of tetracycline and incubation of cells in tetracycline-free medium for 72 h (Fig. 2a). Much to our surprise, none of these three 293T cell lines showed any sign of SAC failure (such as aberrant mitoses or micronucleation; data not shown), although PICH was efficiently depleted (Fig. 2a, b). Furthermore, neither the KT localization of Mad2 nor its levels were detectably affected upon PICH depletion (Fig. 2a, c). Taken at face value, these results suggested the existence of cell-type-specific differences in the response of HeLa and 293T cells to PICH depletion, challenging the idea that PICH is an essential component of the SAC in all cell types.
PICH is not essential for SAC activity in 293T TREX cells. After selection of three different clones (nos. 12, 30, 32) of 293T TREX cells allowing the inducible expression of a PICH-directed shRNA (modeled after PICH-1 siRNA), PICH levels were monitored by Western blotting. In parallel, KT localization of PICH and Mad2 was examined by immunofluorescence microscopy. a Three different clones (12, 30, and 32) of stably transfected 293T TREX-shPICH cells were analyzed by Western blotting for the expression of PICH and Mad2; α-Tubulin was monitored as a loading control. b Bar graph shows quantitative analysis of PICH localization to KTs before and after 72 h tetracycline induction (n > 100). c Bar graph shows quantitative analysis of Mad2 localization to KTs before and after 72 h tetracycline induction (n > 100)
A second disturbing result emerged when we tried to restore SAC activity in PICH-depleted HeLa cells by re-expressing PICH protein at physiological levels. Specifically, we examined the consequences of depleting PICH from a HeLa cell line stably expressing a LAP-tagged mouse homolog of PICH (Poser et al. 2008; Wang et al., manuscript submitted). Although mouse PICH (Ercc6L) shares extensive sequence similarity with its human ortholog (72% amino acid identity; 81% amino acid similarity), expression of LAP-mouse PICH failed to restore SAC functionality in PICH-1 siRNA-treated HeLa cells, as indicated by both extensive micronucleation (Fig. 3a) and accelerated mitosis (Fig. 3b). Yet, LAP-mouse PICH was clearly resistant to PICH-1 siRNA treatment; in fact, LAP-mouse PICH showed enhanced expression upon depletion of endogenous human PICH, suggesting the existence of a feedback mechanism controlling total PICH protein levels (Fig. 3c). These results suggested that either mouse PICH is not able to provide SAC functionality in human cells, that LAP tagging interfered with functionality, or that PICH is not the only target of PICH-1 siRNA.
LAP-mouse PICH fails to rescue PICH-1 siRNA phenotype. a Morphological examination of the nuclei of HeLa cells expressing LAP-mouse PICH after 48 h of siRNA treatment with GL2 or PICH-1 siRNA, respectively. DNA was visualized by DAPI staining. Bar indicates 10 μm. b Mitotic timing in siRNA-treated HeLa cells expressing LAP-mouse PICH. Cells were transfected with the indicated siRNA duplexes and analyzed by live-cell imaging. The bar graph indicates the average time from NEBD to anaphase onset. Data were collected from three experiments (20 cells each), and error bars indicate standard errors. c Cells were transfected with the indicated siRNA duplexes for 48 h and lysates probed by Western blotting with antibodies against PICH, GFP (to selectively visualize LAP-mouse PICH), Mad2, and α-Tubulin
Although earlier observations had suggested that PICH depletion affected Mad2 localization to KTs rather than Mad2 protein levels (Baumann et al. 2007), in the course of the above studies, we noticed that treatment of HeLa cells with the siPICH-1 oligonucleotide caused a reduction in the level of Mad2 protein (Fig. 3a). Thus, the expression of Mad2 protein in response to various PICH siRNA treatments was carefully re-examined. Remarkably, depletion of PICH by all three published PICH-directed siRNAs showed detectable, albeit partial, reduction of Mad2 protein levels (Fig. 4a). Virtually identical results were obtained regardless of whether cells were arrested by thymidine or collected by shake off from an MG132-arrest (data not shown), demonstrating that the observed decrease in Mad2 protein was independent of cell cycle stage. Transfection of cells with siRNAs targeting several genuine SAC components did not cause a reduction of Mad2 protein (Fig. 4b). This rules out general effects related to siRNA transfection per se and demonstrates that the observed reduction of Mad2 levels in PICH siRNA-treated cells is not simply a consequence of SAC abrogation.
Effect of PICH siRNA treatment on Mad2 expression. a HeLa cells were transfected with the indicated siRNA duplexes and levels of PICH, Mad2, and α-Tubulin (loading control) were monitored by Western blotting. b For control, various checkpoint proteins were depleted by siRNA, as indicated, and protein levels determined by Western blotting. c HeLa cells were transfected with the indicated siRNAs and levels of Mad1, Mad2, and PICH mRNA were measured by qRT-PCR. Bar graphs show mRNA levels averaged from three independent experiments (arbitrary units, AU), and error bars represent standard errors
If the previously observed loss of Mad2 protein from KTs in PICH-depleted cells reflected a depletion of Mad2 protein rather than a selective disruption of the Mad1–Mad2 interaction at KTs, we reasoned that it might be possible to restore Mad2 localization to KTs by overexpression of exogenous Mad2 protein in PICH siRNA-treated cells. This was indeed the case, as a Mad2–GFP fusion protein readily localized to KTs in a PICH-1 siRNA background (Supplementary Fig. 2a). In contrast, overexpressed Mad2–GFP did not localize to KTs in cells depleted of Aurora B (Supplementary Fig. 2b), consistent with previous reports (Ditchfield et al. 2003; Vigneron et al. 2004). These observations thus support the conclusion that the PICH siRNA phenotype reported previously might reflect a change in Mad2 abundance rather than localization.
To investigate the mechanism underlying the reduction in Mad2 protein, we next asked whether this reduction was also reflected at the mRNA level. To this end, PICH, Mad1, and Mad2 mRNAs were measured by quantitative real-time polymerase chain reaction (qRT-PCR) in HeLa cells depleted of either PICH or Mad1 for control. As summarized in Fig. 4c, all three PICH siRNAs displayed almost identical knockdown efficiencies with regard to PICH mRNA. Remarkably, all three PICH siRNAs also caused a significant decrease of Mad2 mRNA when compared to the siGL2 control, whereas Mad1 mRNA was not detectably affected (Fig. 4c). As expected, Mad1 siRNA depleted Mad1 mRNA but did not reduce either PICH or Mad2 mRNA (Fig. 4c). Taken together, these results suggest that treatment of HeLa cells with any one of the three previously published PICH siRNA oligonucleotides reduces not only PICH but also Mad2 mRNA levels, suggesting that the previously observed loss of checkpoint functionality might actually reflect a reduction, albeit partial, of cellular Mad2 protein. This in turn implies that either PICH protein somehow regulates Mad2 expression or, alternatively, that the three PICH-directed siRNA oligonucleotides all display off-target effects that cause a lowering of Mad2 expression.
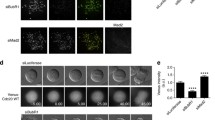
To distinguish between these possibilities, we sought to identify novel siRNA oligonucleotides that would effectively deplete PICH without lowering Mad2 mRNA and protein levels. Indeed, four new siRNAs, referred to as PICH-3, PICH-4, PICH-5, and PICH-6, as well as a corresponding pool (PICH-SP; comprising all four duplexes), were found to deplete PICH effectively. In a direct comparison, PICH-3, PICH-4, and PICH-5 showed comparable efficiency to PICH-1, PICH-2, and PICH-CC siRNA, whereas PICH-6 and the pooled PICH-SP duplexes were slightly less efficient (Fig. 5a). Similar results were obtained regardless of the duration of treatment, indicating that the kinetics of depletion were also comparable (Supplementary Fig. 3). Most importantly and in striking contrast to PICH-1, PICH-2, and PICH-CC, none of the new PICH-directed siRNA oligonucleotides detectably affected Mad2 expression, regardless of whether Mad2 protein (Fig. 5a) or mRNA (Fig. 5b) was monitored. Furthermore, none of the new oligonucleotides detectably interfered with KT localization of Mad2, in contrast to cells treated with PICH-1, PICH-2, and PICH-CC siRNA (Fig. 5c, d). Most importantly, we also examined the influence of the new PICH-directed oligonucleotides on both nocodazole-induced SAC activation (Fig. 5e, f) and the timing of mitotic progression (Fig. 5g). In addition to determining the mitotic index after nocodazole treatment (Fig. 5e), live-cell imaging was performed on HeLa cells stably expressing histone H2B–GFP (Fig. 5f, g). Both mitotic timing (NEBD to anaphase onset) and the duration of the nocodazole-induced SAC arrest were very similar in cells treated with the new PICH-directed siRNAs and in GL2-treated controls, in striking contrast to cells treated with the previously described PICH siRNA oligonucleotides, which advanced anaphase onset and abolished SAC activity (Fig. 5f, g). In our view, the most plausible interpretation of these results is that the originally described PICH-directed siRNA duplexes interfere with SAC activity because they lower Mad2 transcript and protein levels through an off-target effect. We recognize, however, that in principle one could also attribute an off-target effect to those siRNA duplexes that do not lower Mad2 expression. In particular, one could argue that PICH plays a physiological role in controlling Mad2 expression and that the failure of some duplexes to affect Mad2 levels (and hence checkpoint functionality) might reflect off-target effects interfering with this (hypothetical) regulation. A genetic PICH knockout will be required to formally exclude this alternative interpretation.
Analysis of depletion efficiencies and phenotypes produced by different PICH-directed siRNA duplexes. a HeLa cells were treated with indicated siRNAs before lysates were analyzed by Western blotting with the indicated antibodies. Note that Mad2 levels are reduced in response to siPICH-1, siPICH-2, and siPICH-CC (marked by asterisks). b HeLa cells were transfected with the indicated PICH siRNA oligonucleotides and arrested in thymidine prior to RNA purification and measurements of mRNA levels by qRT-PCR. Histogram illustrates relative mRNA levels of Mad1 (blue), Mad2 (red), and PICH (green; in AU). Note that Mad2 mRNA levels are significantly reduced in siPICH-1 and siPICH-CC (marked by asterisk). Error bars indicate standard errors (three independent experiments). c Cells were treated with different PICH-directed siRNAs and stained with antibodies directed against PICH (red) and Mad2 (green); DNA was stained with DAPI (blue). Note that Mad2 is displaced from KTs solely in response to siPICH-1, siPICH-2, or siPICH-CC oligonucleotides. d HeLa cells were treated as in c, and Mad2 mislocalization (displacement from KTs) was counted. The graph shows the percentage of mitotic cells that were depleted of PICH (red bars) as well as the percentage of cells showing mislocalization of Mad2 (blue bars); (n = 50). e Following a 24 h thymidine block, HeLa cells were released for 16 h into nocodazole, and the percentage of mitotic cells was determined by visual inspection after DAPI staining. Graph shows the average from two independent experiments (n = 300). f Histogram summarizes the results of live-cell time-lapse microscopy performed on cells that had been depleted of PICH by 48 h treatments with different PICH-directed siRNA duplexes and then treated with nocodazole during the last 16 h. Error bars indicate standard errors (n = 40). g HeLa cells stably expressing H2B-GFP were transfected with the indicated siRNAs and analyzed by live-cell imaging. The histogram illustrates the elapsed average time from NEBD to anaphase onset (40 cells for each treatment), and error bars indicate standard errors
Re-examination of the role of Tao1 kinase in the spindle checkpoint
Recently, a genome-wide siRNA screen identified a requirement for the Tao1 kinase (also known as microtubule affinity-regulation kinase kinase (MARKK); Johne et al. 2008; Timm et al. 2003) in the spindle checkpoint (Draviam et al. 2007). Moreover, Tao1 depletion was reported to cause a selective loss of Mad2 but not Mad1 from KTs (Draviam et al. 2007), highly reminiscent of the early data obtained after PICH siRNA (Baumann et al. 2007). Intrigued by this similarity, we originally suspected that PICH and Tao1 cooperate in a regulatory step controlling Mad2 localization. However, having obtained evidence that the apparent requirement for PICH in Mad2 localization is likely to reflect an off-target effect, we wondered whether a similar explanation might pertain to Tao1. To explore this possibility, we compared different Tao1-directed siRNA oligonucleotides for their efficiency to deplete Tao1, their effects on Mad2 localization, protein, and transcript levels, as well as their ability to abolish SAC activity. This survey included the most effective previously published Tao1 siRNA oligonucleotide (Tao1-NCB3) as well four new oligonucleotides (Tao1-2 to Tao1-5) and a smart pool comprising Tao1-2 to Tao1-5 (Tao1-SP).
All Tao1-directed siRNAs exhibited a similar ability to deplete Tao1 from HeLa cells, and curiously, PICH-1 siRNA also lowered Tao1 levels (Fig. 6a). Most importantly, siTao1-NCB3 additionally caused a significant reduction of Mad2 protein, comparable to the reduction of Mad2 seen after PICH-1 or PICH-CC siRNA (Fig. 6a). We cannot presently explain the discrepancy between this observation and the previous report, indicating that Tao1 siRNA does not lower Mad2 levels (Draviam et al. 2007). In any case, however, Mad2 levels remained unaffected upon treatment of cells with the other Tao1 siRNAs, except for Tao1-4, which produced a marginal effect (Fig. 6a). Similar results were also obtained upon depletion of Tao1 from U2OS and RPE-1 cells (Supplementary Fig. 4a, b). Quantitative Western blot analysis (three independent experiments; normalized to α-Tubulin) revealed that Tao1-NCB3 siRNA lowered Mad2 protein levels substantially, whereas the other Tao1-directed siRNAs barely affected Mad2, although they depleted Tao1 with similar efficiency as Tao1-NCB3 (Fig. 6b). In line with these observations, the KT localization of Mad2 was abolished only in cells treated with the Tao1-NCB3 siRNA but not in cells treated by any of the other Tao1-directed siRNAs (Fig. 6c, d).
Evaluation of protein knockdown efficiency, Mad2 localization, and SAC phenotype for different siRNA oligonucleotides targeting Tao1 kinase. a HeLa cells were transfected with the indicated siRNA duplexes, and lysates were probed by Western blotting with the indicated antibodies. Asterisks indicate the reduction of Mad2 levels by some but not all Tao1 and PICH-directed siRNAs. b Three independent experiments were performed as in a, and Western blots were used for quantification of Mad2 and Tao1 protein levels. Band intensities were measured using the Aida Image Analyzer software. The histograms show the mean band intensities normalized against background and α-Tubulin, and error bars indicate standard errors. Note that Mad2 protein levels were significantly reduced in lysates from siPICH-1-, siPICH-CC-, and siTao1-NCB3-treated cells (marked by asterisks). c Cells were treated with different Tao1-directed siRNAs and stained with antibodies directed against Mad2 (red); DNA was stained with DAPI (blue). Scale bar indicates 5 µm. Note that Mad2 is displaced from KTs only in response to Tao1-NCB3, but not other Tao1-directed duplexes. d Quantitative analysis of Mad2 localization in cells depleted of Tao1. HeLa cells were treated as described in c. The graph shows the percentage of prometaphase cells in which Mad2 localizes correctly to KTs; shown are the averages from three independent experiments with standard errors (n > 150).
Next, we used qRT-PCR to analyze the mRNA levels of Mad1, Mad2, and Tao1 after treatment of HeLa cells with different siRNAs targeting Tao1 or PICH. While Mad1 mRNA remained constant in all cases, Tao1 mRNA levels were similarly reduced by all Tao1 siRNAs and, to a lesser extent, PICH-1 siRNA (Fig. 7a). Most importantly, Mad2 mRNA was significantly reduced upon treatment with siPICH-1 and siTao1-NCB3 siRNAs and, to a lesser extent, Tao1-4 siRNA. No changes in Mad2 mRNA levels were detectable in cells treated with siPICH-3, siTao1-2, siTao1-3, or control siRNA (siGL2; Fig. 7a). Parallel investigation of cellular phenotypes by time-lapse microscopy (Fig. 7b) revealed that cells depleted of Tao1 by Tao1-NCB3 siRNA went through mitosis more rapidly (NEBD to anaphase; average 14 min) than control cells (average 33 min). Mitotic timing was also reduced in cells treated with the Tao1-4 siRNA, albeit to a lesser extent (average 23 min), but not in response to any of the other Tao1-directed siRNA duplexes (Fig. 7b). Similarly, in line with the original report implicating Tao1 in SAC function (Draviam et al. 2007), an override of the nocodazole-induced SAC arrest could be observed in response to siTao1-NCB3 (average duration of arrest 25 min), whereas a robust checkpoint arrest (often followed by cell death after 12–15 h) was seen upon depletion of Tao1 by siTao1-2 or siTao1-3 (Fig. 7c). Cells treated with Tao1-4 siRNA showed a partial phenotype in that they usually exited mitosis after about 4 h (Fig. 7c). Collectively, we interpret the above data to suggest that the apparent role of the Tao1 kinase in SAC signaling might in reality reflect an off-target effect produced by particular Tao1-directed siRNA oligonucleotides on Mad2, similar to the situation described above for certain PICH siRNA duplexes.
Histogram illustrating mRNA levels, mitotic timing, and nocodazole response after Tao1-directed siRNA. a After treatment of HeLa cells with the indicated siRNAs, RNA was extracted for qRT-PCR measurements. Bars indicate relative mRNA expression levels of Mad1 (blue), Mad2 (red), and Tao1 (green; in AU); shown are averages from three independent experiments and standard errors. Significant reduction of Mad2 mRNA in PICH-1 and Tao1-NCB3 siRNA-treated cells is marked by asterisks. b HeLa cells stably expressing H2B-GFP were transfected with the indicated siRNAs and analyzed by live-cell imaging. The histogram illustrates the elapsed average time from NEBD to anaphase onset (40 cells for each treatment), and error bars indicate standard errors. c Histogram summarizes the results of live-cell time-lapse microscopy performed on cells that had been depleted of Tao1 by 48-h treatments with different Tao1-directed siRNA duplexes and then treated with nocodazole during the last 16 h. Error bars indicate standard errors (n = 40)
Rescue of PICH and Tao1 siRNA phenotypes by Mad2 expression from a bacterial artificial chromosome
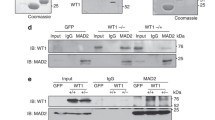
If, as argued above, the abrogation of the SAC in cells treated by some PICH- and Tao1-directed siRNA oligonucleotides were reflecting an off-target effect that causes a reduction of Mad2 levels, it should in principle be possible to rescue this phenotype by re-expressing Mad2. To test this prediction, we used bacterial artificial chromosome transgenesis which is expected to restore protein expression to near physiological levels (Poser et al. 2008). Specifically, a pool of HeLa cells expressing a LAP-tagged murine Mad2 was subjected to PICH, Tao1, and Mad2 siRNA, and SAC functionality was then examined by live-cell imaging (Fig. 8a). In these experiments, GFP-positive cells (expressing murine Mad2) could directly be compared to GFP-negative cells (devoid of murine Mad2). In parallel, the levels of PICH, Tao1, human Mad2, and murine Mad2 were monitored by Western blotting (Fig. 8b). As expected, the mitotic timing (NEBD to anaphase onset) was accelerated in GFP-negative cells subjected to PICH-1, Tao1-NCB3, or Mad2 siRNA, when compared to GL2 controls. In contrast, mitotic timing was similar in GFP-positive cells, regardless of whether they were treated with GL2 control or Mad2 siRNA, attesting to the ability of murine Mad2 to compensate for the depletion of endogenous human Mad2. In the case of PICH siRNA, GFP-positive cells showed a delay in the traverse of mitosis, suggesting that loss of PICH in cells harboring (near) physiological levels of Mad2 actually triggers a mitotic delay. GFP-positive Tao1 siRNA-treated cells also showed a slowing of mitotic traverse compared to GFP-negative cells, although passage through mitosis was still faster than in GL2-treated controls. Taken at face value, this would be consistent with the notion that Tao1 is required for full SAC functionality even in the presence of physiological levels of Mad2 (Draviam et al. 2007). However, we note that all GFP-positive, murine Mad2-expressing cells showed a prolonged arrest in response to nocodazole, regardless of whether they were treated with PICH, Tao1, or (human) Mad2 siRNA (data not shown). Collectively, these data make a strong case that the observed reduction of Mad2 levels is largely, although in the case of Tao1 perhaps not exclusively, responsible for the abrogation of SAC activity in the PICH- and Tao1-siRNA-treated cells.
Rescue of PICH-1 and Tao1 siRNA phenotypes by LAP-mouse Mad2. a Mitotic timing in siRNA-treated HeLa cells expressing LAP-mouse Mad2. Cells were transfected for 48 h with the indicated siRNA duplexes and analyzed by live-cell imaging. In these experiments, GFP-positive cells (expressing mouse Mad2) and GFP-negative cells (not expressing mouse Mad2) were monitored in parallel. The bar graph indicates the average time from NEBD to anaphase onset. Data were collected from four experiments (20–25 cells each), and error bars indicate standard errors. b Lysates were prepared from cells treated as in a and analyzed by Western blotting with the indicated antibodies. The lane to the right represents a lysate prepared from the parental HeLa (Kyoto) cell line that lacks the BAC expressing murine Mad2. Asterisk denotes a cross-reactive band that serves as loading control
Uncovering of a regulatory influence of Plk1 on Mad2 function
A critical role for PICH and Tao1 in the SAC had originally been supported not only by consistent siRNA phenotypes but also apparently successful rescue experiments (this study, Fig. 1; Draviam et al. 2007). In particular, the SAC abrogation produced by Tao1-NCB3 siRNA could successfully be rescued by active but not inactive kinase, strongly supporting a role for Tao1 in the SAC (Draviam et al. 2007). However, the published evidence suggests that this rescue involved overexpression of siRNA-resistant Tao1 (Draviam et al. 2007), raising the possibility of a “bypass suppression” (in genetic terms) rather than a genuine rescue. In particular, we consider it possible that excess exogenous Tao1 activity might have interfered with chromosome attachment to the spindle apparatus in such a way that even reduced levels of Mad2 might have been sufficient to restore an SAC response. So it would seem critical to demonstrate restoration of SAC activity in Tao1-depleted cells by physiological levels of Tao1 kinase.In the case of PICH, our results clearly demonstrate that overexpression of PICH protein restored SAC functionality in PICH-1 siRNA-treated cells (Fig. 1), and yet, physiological levels of LAP-mouse PICH provided no rescue (Fig. 3b). A first clue for how to explain this discrepancy emerged when we tested two PICH mutants for their ability to restore normal mitotic timing in PICH-1 siRNA-treated cells. In fact, a PICH protein carrying a mutation predicted to abolish its ATPase activity (K128A, Wang et al., submitted; Leng et al. 2008) readily restored SAC activity in PICH-1 siRNA-treated cells, whereas a mutant that cannot bind Plk1, due to a mutation within the so-called Polo-box domain (T1063A), failed to restore proper mitotic timing (Fig. 9a). These unexpected results could not be attributed to different expression levels of the rescuing constructs, as neither the transfection efficiencies nor the levels of overexpression achieved revealed significant differences (Fig. 9b). Instead, they demonstrate that restoration of SAC functionality in PICH-1 siRNA-treated cells depends on the ability of the overexpressed PICH protein to sequester Plk1, rather than its ATPase activity. One straightforward interpretation of this result is that residual Mad2 protein, as it persists after PICH-1 siRNA, was sufficient to provide SAC activity, provided that Plk1 was sequestered by exogenous wild-type or ATPase-deficient PICH protein. Thus, the observed restoration of SAC activity would represent a case of bypass suppression rather than genuine rescue.
Analysis of overexpressed PICH mutants for their ability to restore mitotic timing in PICH-1 siRNA-treated cells. a mCherry-PICH WT (shown in Fig. 1), mCherry-PICH-K128A (defective in ATPase activity), and mCherry-PICH-T1063A were cotransfected with PICH-1 siRNA oligonucleotide into H2B-GFP-expressing HeLa cells before these were analyzed by live-cell imaging (as described in the legend to Fig. 1). mCherry-positive cells were analyzed to determine the average time from NEBD to anaphase onset. Left panels show single-plane images selected from movies and time stamps indicate hours:minutes. Right panels show representative fate maps of 25 individual cells (the whole experiment being repeated independently three times), with red columns indicating the time span from NEBD to anaphase onset and blue columns indicating cells that died in interphase during imaging. Average times from NEBD to anaphase onset are indicated to the right. b Quantification of overexpression of mCherry-PICH WT, mCherry-PICH K128A, and mCherry-PICH T1063A in comparison to endogenous PICH. HeLa cells were transfected with the indicated plasmids as in a, and total cell lysates were subjected to serial dilutions before Western blotting with anti-PICH antibody. Transfection efficiency (indicated to the right) was determined by immunofluorescence staining using anti-mCherry antibody, and fold overexpression was calculated, taking into account the dilution factor and the transfection efficiency. This experiment indicates that the mCherry-PICH constructs were similarly overexpressed (about 20–24-fold) when compared to endogenous PICH levels
A corollary of the above interpretation is that Plk1 activity contributes to regulate the efficacy of Mad2 in SAC signaling, so that less Mad2 is needed for SAC functionality in the absence of Plk1 than in its presence. If this were the case, one would predict that siRNA-mediated depletion of Plk1 or inhibition of Plk1 by the small molecule inhibitor TAL (Santamaria et al. 2007) should also be able to restore SAC activity in PICH-1 siRNA-treated cells. To directly test this prediction, we performed live-cell imaging on HeLa cells stably expressing histone H2B–GFP and monitored mitotic progression under various treatments. Cells were depleted of either PICH or Tao1, using PICH-1 or Tao1-NCB3 siRNA, respectively, and some samples were subjected to additional depletion of Plk1 by siRNA or treated with TAL. Whereas cells treated with PICH-1 siRNA alone showed no mitotic arrest in response to nocodazole (Fig. 5f), codepletion of Plk1 or inhibition of Plk1 by TAL resulted in mitotic delays of 120 and 160 min, respectively (Fig. 10a), before cells exited mitosis without chromosome segregation and often formed micronuclei (data not shown). Single depletion of Plk1 or inhibition of Plk1 caused a prolonged prometaphase delay, mostly followed by apoptosis (Fig. 10a), as expected (Santamaria et al. 2007; Liu and Erikson 2003; Petronczki et al. 2008).
Codepletion of PICH and Plk1 restores SAC activity. a–c HeLa cells stably expressing H2B-GFP were subjected to single and double depletion (PICH, Tao1, Mad2, Plk1) or inhibition (Plk1) experiments, as described in each panel, and analyzed by time-lapse microscopy. Histograms illustrate the mitotic timing from NEBD to mitotic exit or cell death. Error bars represent standard error (n = 40). d Western blot analysis of Mad2 abundance after transfection of HeLa cells with increasing concentrations of Mad2 siRNA duplex (0.06, 1.2, 3.2, 6.6, 20 pM) and PICH-1, Tao1-NCB3, and GL2 siRNA for comparison. α-Tubulin serves as loading control. e HeLa cells stably expressing H2B-GFP were transfected with increasing amounts of Mad2 siRNA (as described in d) as well as PICH-1 and Tao1-NCB3 duplexes, prior to synchronization with thymidine and release into 1 µM TAL-containing medium. Then, the mitotic timing from NEBD to anaphase onset was determined by live-cell imaging. Histograms illustrate the results from three independent experiments (n = 20 for each experiment), and error bars represent standard error
We also analyzed the effect of Plk1 depletion or inhibition on cells that were treated with Tao1-NCB3 siRNA (Fig. 10b). Whereas control cells exhibited the expected mitotic timing, Tao1-NCB3 siRNA-treated cells separated their chromosomes in less than 20 min without forming a metaphase plate. The simultaneous depletion of Plk1 by siRNA or its inhibition by TAL increased the timing in Tao1-NCB3 siRNA-treated cells to about 35 min, similar to the duration seen in control cells but far below the duration of the mitotic arrest seen after depletion of Plk1 alone (Fig. 10b). This suggests that Plk1 depletion or inhibition only marginally restored SAC activity in Tao1-NCB3 siRNA-treated cells, presumably because residual Mad2 levels were too low (see below).
To demonstrate that residual Mad2 was critical for the restoration of SAC activity by Plk1 depletion/inhibition in PICH-1 or Tao1-NCB3 siRNA-treated cells, we also examined the consequences of combining Plk1 inhibition with direct siRNA-mediated depletion of Mad2. As shown in Fig. 10c, TAL addition could not restore normal mitotic timing to cells that were extensively depleted of Mad2, arguing that the absence of Plk1 activity cannot rescue SAC functionality in cells from which the bulk of Mad2 has been depleted. These results support the view that residual Mad2, as it persists after PICH-1 or Tao1-NCB3 siRNA treatment, is critical for the restoration of some SAC activity upon depletion or inhibition of Plk1. As a final test of this conclusion, different amounts of Mad2 siRNA oligonucleotides were used to reduce cellular Mad2 to levels within the range of those seen as a consequence of PICH-1 or Tao1-NCB3 siRNA treatment (Fig. 10d). Then, live-cell imaging was used to analyze mitotic timing in TAL-treated cells as a function of Mad2 levels. As shown in Fig. 10e, the mitotic timing (NEBD to anaphase onset) observed in response to Plk1 inhibition showed a strong correlation with Mad2 levels. Furthermore, Tao1-NCB3 siRNA resulted in a more severe depletion of Mad2 than PICH-1 siRNA and (Fig. 10d), concomitantly, a more pronounced advancement of mitotic timing (Fig. 10e), in excellent agreement with the data shown above (Fig. 10a, b). On the basis of these results, we propose that restoration of SAC activity by depletion or inhibition of Plk1 depends on residual levels of Mad2, implying that depletion or inhibition of Plk1 enhances the functionality of Mad2. In turn, these observations point to the conclusion that—under physiological circumstances—Plk1 antagonizes that SAC signaling function of Mad2.
Conclusions
Several conclusions emerge from this study. First and foremost, our results question the previously reported requirements for PICH (Baumann et al. 2007) and Tao1 (Draviam et al. 2007) for SAC activity. In the case of PICH, we find it difficult to escape the conclusion that the previous siRNA-based implication of this protein in the SAC reflects an off-target effect on Mad2. At present, the precise physiological function of PICH awaits further study, but considering its conspicuous localization to centromeres, KTs, and UFBs, a role in chromosome structure and segregation appears likely. This view is supported by the physical interaction between PICH and Plk1 and the influence the two proteins exert on each other’s localization (Baumann et al. 2007; Santamaria et al. 2007; Leng et al. 2008). Our study on Tao1 is less extensive, and some of our results differ from those reported in the original study, notably with regard to the effect of siRNA on Mad2 and the advancement of mitotic timing (Draviam et al. 2007). Also, we emphasize that Draviam and coworkers reported an effective rescue of Tao1 depletion by active but not inactive kinase (Draviam et al. 2007). Thus, further study will be required to definitively validate or exclude a role for Tao1/MARKK in the SAC. In any case, considering the role of this kinase in the regulation of microtubules (Johne et al. 2008; Timm et al. 2003), an important contribution to mitotic events is to be expected.
To our surprise, we discovered that Mad2 levels are frequently reduced as a consequence of off-target effects exerted by siRNA oligonucleotides. Considering that the alignment of the Mad2 mRNA sequence with the sequences of the various PICH- or Tao1-directed oligonucleotides revealed no evidence for extensive complementarities (Fig. 11 and data not shown), the Mad2 transcript is unlikely to be the primary target of these siRNA duplexes. Instead, we presume that the PICH-1, PICH-2, PICH-CC, and Tao1-NCB3 duplexes target other transcripts, as yet unidentified, which then affect Mad2 mRNA levels through indirect mechanism(s). Microarray experiments and/or deep sequencing will hopefully shed light on the target range of these siRNA oligonucleotides. In turn, the identification of critical targets may contribute to a better understanding of the mechanisms that regulate Mad2 expression. From a broader perspective, an important question is why Mad2 appears to be such a sensitive, albeit unintentional, target of siRNA experiments. We see at least three plausible explanations, which are not mutually exclusive. First, although many proteins (including SAC components) provide functionality even at reduced levels, this is clearly not the case for Mad2, whose exact levels are critical for function (Michel et al. 2001; Hernando et al. 2004). Therefore, even partial reductions in Mad2 levels are expected to produce pronounced phenotypes. Second, Mad2 plays a central role in SAC signaling and so its expression is likely to be regulated by multiple mechanisms. This in turn provides a broad range of transcripts and pathways that could potentially be modulated by small RNAs, perhaps including siRNAs that function as microRNAs. Third, the loss of SAC activity that accompanies the lowering of Mad2 levels is relatively easy to score in cellular assays. Thus, a general prediction emerging from the present study is that off-target effects will be identified more readily if they converge onto proteins that are critically involved in cellular processes that can easily be monitored.
Alignment of PICH and Tao1 siRNA sequences with Mad2 mRNA. The alignments were performed using the global EMBOSS pairwise Alignment Algorithm (http://www.ebi.ac.uk/Tools/emboss/align) with a gap open of 50.0, and the Mad2 RNA accession number NM_002358.3. The most extensive match observed for each oligonucleotide is shown. Alignments colored in red or green refer to siRNA duplexes that do or do not lower Mad2 transcripts, respectively
Finally, our study illustrates that even rescue experiments, the accepted “gold standard” for assessing siRNA specificity, must be interpreted with caution, unless rescuing proteins are re-expressed at physiological levels. In this study, we have in fact discovered that the apparent “rescue” of a siRNA phenotype depended on the ability of the “rescuing” PICH protein to sequester the mitotic kinase Plk1. This intriguing case of bypass suppression suggests that Plk1 normally reduces the strength of the Mad2-dependent inhibitory SAC signal. In future studies, it will clearly be interesting to explore the molecular mechanism(s) by which Plk1 affects SAC functionality.
Materials and methods
Cell lines
HeLa, U2OS, and HeLa cells stably expressing H2B–GFP were grown under standard conditions in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. Cell cycle synchronizations were performed by a single thymidine (2 mM) block 10–12 h posttransfection with siRNA duplexes. For microscopic analysis of mitotic cell cycle stages, cells were released from thymidine for 12 h before fixation. Mitotic cells were isolated by mitotic shake off. The stable 293T TREX-shPICH cell lines were prepared under selective conditions using blasticidin (5 µg/ml) and puromycin (1 µg/ml) and grown under standard conditions. Stable clones were picked and analyzed by Western blotting and immunofluorescence microscopy before and after tetracycline induction.
HeLa cells (pools) containing bacterial artificial chromosomes (BACs) expressing C-terminally LAP-tagged mouse PICH (Ercc6L, BAC-ID RP24-326A8) or mouse Mad2L1 (BAC-ID RP23-84G11) were produced as described previously (Poser et al. 2008). In brief, BACs were obtained from the BACPAC Resources Center (http://bacpac.chori.org), the LAP (EGFP-IRES-Neo)-cassette was amplified by PCR using primers that carry homology to the C terminus of Ercc6L and Mad2L1, respectively, prior to transfection of the modified BAC.
Transfection of plasmids, siRNA, and plasmid constructions
Plasmid and siRNA transfections were performed using TransIT-LT1 transfection reagent (Mirus) and Oligofectamine reagent (Invitrogen), respectively, according to the manufacturer’s instructions. The siRNA sequences of GL2 (Elbashir et al. 2001), Mad2, PICH-1, PICH-2 (Baumann et al. 2007), PICH-CC (Leng et al. 2008), Tao1-NCB3 (Draviam et al. 2007), Aurora B (Klein et al. 2006), Mps1, and Mad1 (Stucke et al. 2002) have been published previously. The following siRNA target sequences were purchased from Dharmacon (ON-TARGETplus) unless otherwise stated: PICH-3: 3′-AGUAGGUGGUGUCGGUUUA; PICH-4: 3′-GGAUAGAGUUUACC GAAUU; PICH-5: 3′-CCAGAAACCUCAAUCGGAU; PICH-6: 3′-ACUUUAAGACAUUGCGAAU; the PICH oligo SMARTpool contained a mixture of PICH-3, PICH-4, PICH-5, and PICH-6 siRNAs; Tao1-2: 3′-CCAAGUAUCUCGUCACAAA; Tao1-3: 3′-GUAAUAUGGUCCUUU CUAA; Tao1-4: 3′-CUAAAGUGAUGUCCAAUGA; Tao1-5: 3′-GCUGUGAGUUGAUCAGAUU; the Tao1 oligo SMART pool contained a mixture of Tao1-2, Tao1-3, Tao1-4, and Tao1-5 siRNAs; Bub1: 3′-CCAGTGAGTTCCTATCCAATT (Qiagen).
To clone PICH constructs for expression in HeLa cells, PICH cDNA was amplified by PCR using pEGFP-C1-PICH (WT and T1063A; Baumann et al. 2007) as a template and the following primers: M3863-TCTCCCCGGGATGGAGGCATCCCGAAGGTTTC and M3864-ATAAGAATGCGGCCGCTCAATTGTTATTAAGTTGC. The Walker A mutant (K128A) was generated by site-directed mutagenesis using the following two oligonucleotides: M3821-GATGATATGGGATTAGGGGCGACTGTTCAAATCATTGCT and M3822-AGCAATGATTTGAACAGTCAGTCGCCCCTAATCCCATATCATC. For rescue experiments, mCherry-tagged PICH constructs were used. The cDNA for mCherry was PCR-amplified from pRSET-B-mCherry (a kind gift from Roger Y Tsien), using the following primers: M3914-CGGGGTACCGCCACCATGGTGAGCAAGGGCGAGGAGGAT and M3915-GCGATATCCTTGTACAGCTCGTCCATGCCG. The mCherry construct was then subcloned into KpnI and EcoRV sites of the pcDNA4/TO vector. Both WT and mutant PICH cDNAs were subcloned into the pcDNA4/TO-mCherry vector. To make constructs resistant against PICH-1 siRNA (Baumann et al. 2007), the above constructs were then subjected to site-directed mutagenesis using the following primers: M3344-GAGGGTGAGAAACAAGACTTATCCAGTATAAAGGTG and M3345-CACCTTTATACTGGATAAGTCTTGTTTCTCACCCTC. For construction of the shPICH plasmid, the following oligonucleotides were annealed in annealing buffer (200 mM Tris pH 7.5, 100 mM MgCl2 and 500 mM NaCl) for 1 h at 37°C and cloned into pTER +vector using BglII and HindIII restriction sites: M3666-GATCCCCAAGATCTCTCCAGTATA TTCAAGAGATTATACTGGAGAGATCTTGTTTTTGGAAA and M3667-AGCTTTTCCAAAAACAAGATCTCTCCAGTATAATCTCTTGAATTATACTGGAGAGATCTTGGG. (Italic letters indicate the siRNA sequence published for siPICH-1 (Baumann et al. 2007)). Full-length Mad2 was amplified by PCR and subcloned into pEGFP-C2 through BamH1 and Sal1 restriction sites. All constructs were verified by sequencing.
Live-cell imaging
For imaging experiments, HeLa cells stably expressing histone H2B–GFP were used (Sillje et al. 2006). They were cultured in eight-well chamber slides (Ibidi) with 300 μl medium per well and transfected with siRNA oligonucleotides and/or plasmid constructs, as appropriate. Ten hours posttransfection, they were synchronized using 2 mM thymidine for 24 h and then released into fresh medium for 8 h prior to imaging. Live-cell imaging was performed using a Zeiss Axiovert 2 microscope equipped with a Plan Neofluar 20x1.6 optivar/NA = 0.75 objective, an environmental chamber, and a CoolSNAP-ES2 digital camera system. Movies were acquired over at least 12 h, with pictures taken at time intervals of 3 min. Images were captured with 2.5–5 ms exposure time for histone H2B–GFP, 30 ms for mCherry and 50 ms for DIC. Metaview software (Visitron Systems) was used for data collection and analysis.
Western blotting
Whole cell extracts were prepared using lysis buffer (20 mM Tris/HCl pH 7.4, 150 mM NaCl, 40 mM β-glycerophosphate, 10 mM NaF, 0.5% IGEPAL, 2 mM Pefabloc (Roth), 0.3 mM NaVO3, 100 µM ATP, 100 µM MgCl2, 100 nM okadaic acid, one protease inhibitor tablet per 10 ml lysis buffer (Roche)). Proteins were resolved by sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore), and these were probed with primary antibodies overnight at 4°C. Membranes were washed with 0.5% Tween/phosphate-buffered saline (PBS) before incubation with secondary antibodies. Antibodies used were rabbit anti-Mad2 (Bethyl), rabbit anti-Tao1 (Bethyl), rabbit anti-PICH (Baumann et al. 2007), monoclonal anti-α-Tubulin (DM1A, Sigma), mouse anti-Mad1 (Sigma), mouse anti-Aurora B (AIM1; BD Transduction), monoclonal anti-Mps1 (Stucke et al. 2002). Monoclonal anti-Bub1 and monoclonal anti-GFP antibodies were kindly provided by Andreas Uldschmidt.
Immunofluorescence microscopy
Cells were grown on coverslips, fixed with PTEMF for 10 min at room temperature (Stucke et al. 2002), and incubated for 20 min at room temperature in 3% bovine serum albumin/PBS. Antibody incubations were carried out for 1 h at room temperature, followed by three washes in 0.5% TritonX-100/PBS. DNA was stained with 1 µg/ml 4,6-diamidino-2-phenylindole (DAPI). For image acquisition, a Deltavision microscope (Applied Precision) on a Nikon Eclipse TE200 base (Applied Precision) equipped with an Apo 60/1.4 oil immersion objective and a CoolSnap HQ camera (Photometrics) was used. Samples were examined with optical sections acquired 0.4 µm apart in the z-axis, deconvolved for each focal plane and projected into a single-plane image using the Softworx software (Applied Precision). For immunostaining, rabbit anti-Mad2 (Bethyl), rat anti-PICH (Baumann et al. 2007), and mouse anti-Aurora B (AIM1; BD Transduction) were used. Primary antibodies were detected by Cy3-conjugated donkey antibodies (Dianova) and Alexa-Fluor-647-conjugated goat antibodies (Invitrogen).
Quantitative real-time PCR
To monitor mRNA levels, HeLa cells were depleted of various proteins by siRNA and synchronized by a single thymidine block. To analyze the expression levels of Mad1, Mad2, PICH, and Tao1 transcripts, total RNA was extracted using an RNeasy Mini kit according to manufacturer’s protocol (Qiagen) and quality-controlled for integrity by capillary electrophoresis on Agilent 2100 Bioanalyzer. Transcript levels were determined by qRT-PCR. First-strand cDNA synthesis was carried out from 1 µg of total RNA by using SuperScript II Reverse Transcriptase and random primers following the manufacturer’s instructions (Invitrogen Life Technologies). The amplicons were designed with the program Primer Express 2.0 (Applied Biosystems, Foster City, CA, USA) by using default parameters such that they spanned exon boundaries. Primer sequences are available upon request. Amplicon sequences where checked by BLAST against the human genome to ensure that they were specific for the gene being assayed. The specificity of each primer pair as well as the efficiency of the amplification step was tested by assaying serial dilutions of cDNA. PCR reactions were carried out in triplicate by using an SDS 7900 HT instrument (Applied Biosystems) and Power SYBR Green PCR master mix kits (Applied Biosystems). Normalization genes were selected using the geNorm script as published (Vandesompele et al. 2002). Raw Ct values obtained with SDS 2.0 (Applied Biosystems) were imported into Excel (Microsoft, Redmond, WA, USA) to calculate the normalization factor and the fold changes with the geNorm (Vandesompele et al. 2002).
References
Bachrati CZ, Hickson ID (2008) RecQ helicases: guardian angels of the DNA replication fork. Chromosoma 117(3):219–233
Baumann C, Korner R, Hofmann K, Nigg EA (2007) PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 128(1):101–114
Chan KL, North PS, Hickson ID (2007) BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. Embo J 26(14):3397–3409
Chan KL, Palmai-Pallag T, Ying S, Hickson ID (2009) Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11(6):753–760
Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS (2003) Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol 161(2):267–280
Draviam VM, Stegmeier F, Nalepa G, Sowa ME, Chen J, Liang A, Hannon GJ, Sorger PK, Harper JW, Elledge SJ (2007) A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling. Nat Cell Biol 9(5):556–564
Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411(6836):494–498
Hauf S, Waizenegger IC, Peters JM (2001) Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293(5533):1320–1323
Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, Lowe SW, Cordon-Cardo C (2004) Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 430(7001):797–802
Johne C, Matenia D, Li XY, Timm T, Balusamy K, Mandelkow EM (2008) Spred1 and TESK1—two new interaction partners of the kinase MARKK/TAO1 that link the microtubule and actin cytoskeleton. Mol Biol Cell 19(4):1391–1403
Klein UR, Nigg EA, Gruneberg U (2006) Centromere targeting of the chromosomal passenger complex requires a ternary subcomplex of borealin, survivin, and the N-terminal domain of INCENP. Mol Biol Cell 17(6):2547–2558
Leng M, Bessuso D, Jung SY, Wang Y, Qin J (2008) Targeting Plk1 to chromosome arms and regulating chromosome compaction by the PICH ATPase. Cell Cycle 7(10):1480–1489
Liu X, Erikson RL (2003) Polo-like kinase (Plk)1 depletion induces apoptosis in cancer cells. Proc Natl Acad Sci U S A 100(10):5789–5794
Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VV, Benezra R (2001) MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 409(6818):355–359
Naim V, Rosselli F (2009) The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol 11(6):761–768
Petronczki M, Lenart P, Peters JM (2008) Polo on the rise—from mitotic entry to cytokinesis with Plk1. Dev Cell 14(5):646–659
Poser I, Sarov M, Hutchins JR, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, Pelletier L, Kittler R, Hua S, Naumann R, Augsburg M, Sykora MM, Hofemeister H, Zhang Y, Nasmyth K, White KP, Dietzel S, Mechtler K, Durbin R, Stewart AF, Peters JM, Buchholz F, Hyman AA (2008) BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods 5(5):409–415
Santamaria A, Neef R, Eberspacher U, Eis K, Husemann M, Mumberg D, Prechtl S, Schulze V, Siemeister G, Wortmann L, Barr FA, Nigg EA (2007) Use of the novel Plk1 inhibitor ZK-thiazolidinone to elucidate functions of Plk1 in early and late stages of mitosis. Mol Biol Cell 18(10):4024–4036
Sillje HH, Nagel S, Korner R, Nigg EA (2006) HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol 16(8):731–742
Stucke VM, Sillje HH, Arnaud L, Nigg EA (2002) Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. Embo J 21(7):1723–1732
Timm T, Li XY, Biernat J, Jiao J, Mandelkow E, Vandekerckhove J, Mandelkow EM (2003) MARKK, a Ste20-like kinase, activates the polarity-inducing kinase MARK/PAR-1. Embo J 22(19):5090–5101
Uhlmann F, Lottspeich F, Nasmyth K (1999) Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400(6739):37–42
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):RESEARCH0034
Vigneron S, Prieto S, Bernis C, Labbe JC, Castro A, Lorca T (2004) Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol Biol Cell 15(10):4584–4596
Wang LH, Schwarzbraun T, Speicher MR, Nigg EA (2008) Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma 117(2):123–135
Acknowledgements
We thank Roger Tsien and Andreas Uldschmid for reagents, Christelle Barraclough for help with qRT-PCR, and Rainer Malik for help with bioinformatics. We also thank Christoph Baumann, Viji Draviam, Steve Elledge, and all members of the Nigg lab for helpful discussions. This work was supported by the Max-Planck-Society. L.H. W. acknowledges a fellowship from the Taiwan Merit Scholarships program (NSC-095-SAF-I-564-627-TMS). NCH gratefully acknowledges the Ph.D. fellowship from the Boehringer Ingelheim Fonds.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Musacchio
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Cells treated with PICH siRNA oligonucleotides display defective SAC. HeLa cells stably expressing histone H2B–GFP were treated with three different siRNA duplexes targeting PICH (siPICH-1, siPICH-2, and siPICH-CC), synchronized by a single thymidine block and analyzed by time-lapse microscopy 48 h after transfection. a Selected frames from representative live-cell movies show cells that traverse mitosis after treatment with control (GL2) or PICH-directed siRNA. Upper panels show GFP fluorescence; lower panels show differential interference contrast (DIC). Note that cells lacking PICH initiate anaphase without awaiting metaphase plate formation. b Cells were treated with nocodazole and filmed as described in a. Note that PICH-depleted cells fail to sustain the nocodazole-induced checkpoint arrest that is seen in control siRNA (GL2)-treated cells. All time stamps represent hours:minutes (GIF 242 kb)
Supplementary Fig. 2
Overexpressed Mad2-GFP localizes to KTs in PICH-depleted cells. a, b Immunofluorescence microscopy images of prometaphase cells after treatment with the indicated siRNAs (48 h) and transfection (during the last 24 h) with either empty-GFP vector or GFP-tagged Mad2. Cells were fixed with PTEMF and costained with PICH (a) or Aurora B (b) antibody (red), respectively. DNA was stained with DAPI (blue). Note that overexpressed Mad2–GFP localizes to KTs even in the absence of PICH. Scale bars represent 5 µm (GIF 278 kb)
Supplementary Fig. 3
All PICH siRNA duplexes deplete PICH with similar depletion efficiency and kinetics. HeLa cells were depleted of PICH by six different siRNA duplexes and one smart pool for 24, 48, and 72 h and probed by Western blotting with the indicated antibodies; α-tubulin serves as loading control (GIF 84.6 kb)
Supplementary Fig. 4
Effects on Mad2 protein levels after depletion of PICH and Tao1 from U2OS and RPE-1 cells. U2OS (a) and RPE-1 cells (b) were treated with the indicated PICH- and Tao1-directed siRNA duplexes for 48 h, and the abundance of both the primary targets and Mad2 protein was analyzed by Western blotting. The asterisks indicate significant reduction of Mad2 protein levels (GIF 79.4 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Hübner, N.C., Wang, L.HC., Kaulich, M. et al. Re-examination of siRNA specificity questions role of PICH and Tao1 in the spindle checkpoint and identifies Mad2 as a sensitive target for small RNAs. Chromosoma 119, 149–165 (2010). https://doi.org/10.1007/s00412-009-0244-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-009-0244-2