Abstract
Small nuclear ribonucleoproteins (snRNPs) play a fundamental role in pre-mRNA processing in the nucleus. The biogenesis of snRNPs involves a sequence of events that occurs in both the nucleus and cytoplasm. Despite the wealth of biochemical information about the cytoplasmic assembly of snRNPs, little is known about the spatial organization of snRNPs in the cytoplasm. In the cytoplasm of larch microsporocytes, a cyclic appearance of bodies containing small nuclear RNA (snRNA) and Sm proteins was observed during anther meiosis. We observed a correlation between the occurrence of cytoplasmic snRNP bodies, the levels of Sm proteins, and the dynamic formation of Cajal bodies. Larch microsporocytes were used for these studies. This model is characterized by natural fluctuations in the level of RNA metabolism, in which periods of high transcriptional activity are separated from periods of low transcriptional activity. In designing experiments, the authors considered the differences between the nuclear and cytoplasmic phases of snRNP maturation and generated a hypothesis about the direct participation of Sm proteins in a molecular switch triggering the formation of Cajal bodies.
Similar content being viewed by others
Introduction
Eukaryote nuclei contain dynamic structures called Cajal bodies. Cajal first observed these nuclear bodies in rat neurons stained with silver salts using the Ag-NOR technique (Cajal 1903). By using electron microscopy, the Cajal bodies were revealed to consist of coiled fibrils that resemble a coil of twisted thread. For this reason, these nuclear domains have frequently been called “coiled bodies” (CB, Monneron and Bernhard 1969). Until the 1970s and 1980s, it was not known that Cajal bodies are also present in plants (Górska-Brylass and Wróbel 1978; Moreno Díaz de la Espina et al. 1982). Currently, we know that CBs are evolutionarily conserved structures, which are found in the majority of animal and plant cell types (for review, see Shaw and Brown 2004; Cioce and Lamond 2005). Recently, plant Cajal bodies have been shown to also contain Atcoilin (Collier et al. 2006; Koroleva et al. 2009), which is a homolog of the CB marker protein coilin in animal cells. This finding suggests that CBs have a basic role in nuclear function in all eukaryotic cells; however, Cajal bodies in plants have been implicated as sites of siRNA biogenesis, whereas in animals siRNA dicing occurs in the cytoplasm (for review, see Pontes and Pikaard 2008). Recently, we have demonstrated the presence of poly(A) RNA in Cajal bodies (Kołowerzo et al. 2009). Such poly(A) RNA localization had not yet been observed in animal cells (Visa et al. 1993; Huang et al. 1994). Further study (Smoliński and Kołowerzo 2011) demonstrated the presence of several housekeeping gene transcripts in CBs. While CBs in all organisms contain components of the pre-mRNA splicing machinery, other machineries vary in CB localization. In contrast to the amphibian CBs (Wu and Gall 1993) and in HeLa cell nuclei CBs (Frey and Matera 1995), recent studies in Drosophila showed that the U7 snRNP is not concentrated in CBs but in a separate structure, the histone locus body (HLB) (Liu et al. 2006b).
One characteristic trait of Cajal bodies is the dynamic changes in their size and number in different species, individual tissues, and cells (Boudonck et al. 1998; Acevedo et al. 2002). Moreover, it has been shown that the number of CBs in plant cells changes during the cell cycle and differentiation (Boudonck et al. 1998; Straatman and Schel 2001; Seguí-Simarro et al. 2006; Zienkiewicz and Bednarska 2009).
It has been experimentally established that the number of Cajal bodies is related to the metabolic activity of a cell. A positive correlation between the formation of Cajal bodies and transcriptional activity was revealed in HeLa cells (Ferreira et al. 1994), in hamster embryos (Ferreira and Carmo-Fonseca 1996), in oocytes (Chouinard 1975; Parfenov et al. 2003) and during microspore embryogenic development (Seguí-Simarro et al. 2006), and pollen development (Zienkiewicz and Bednarska 2009) in plant cells. However, there are exceptions. In metabolically active cells like single-cell mouse embryos, CBs appear 16 h after fertilization, preceding the initiation of transcription that usually occurs in two-cell embryos (Ferreira and Carmo-Fonseca 1995). Cajal bodies are present in insect and mammalian oocytes, despite chromatin condensation in partially or completely inactive nuclei (Bogolyubov and Parfenov 2001; Parfenov et al. 2003; Batalova et al. 2005). Also a prominent Cajal body (endobody) was present during most of prophase, attached to the karyosome (Liu et al. 2006a) in the germinal vesicle of the Drosophila oocyte transcriptionally quiescent during the latter part of the first meiotic prophase. Moreover, an increase in the number of CBs was found in cells of hibernating dormice (Malatesta et al. 1994). The visualization of CBs in living plant cells suggests that significant changes in the number of Cajal bodies may be related to their ability to fuse to form larger CBs or divide into smaller ones (Boudonck et al. 1999).
Another strategy that cells use to increase the number of Cajal bodies is de novo formation. This complex process is strictly correlated with the small nuclear ribonucleoproteins (snRNPs) cycle (Navascues et al. 2004). snRNP biogenesis includes a sequence of events that occur in both the nucleus and cytoplasm. The first stage of this cycle is the synthesis of small nuclear RNA (snRNA) molecules in the nucleus. The U1, U2, U4, and U5 molecules are transcribed by polymerase II and acquire a 7-methylguanosine (m7G) cap at the 5′ end and additional nucleotides at the 3′ end as a result of the co-transcription process. The cap at the 5′ end is recognized by the cap-binding complex (CBC). This complex is necessary to transport the snRNAs to the cytoplasm. The PHAX protein (phosphorylated adapter for RNA export) is the second factor that promotes the export of snRNPs from the nucleus (Navascues et al. 2004). First, the binding of Sm proteins to the snRNA and the maturation of the 3′ end occur in the cytoplasm. Next, the snRNA acquires a 2,2,7-methylguanosine (m3G) cap at the 5′ end (Mattaj 1986; Dickmanns and Ficner 2005).
Despite the wealth of biochemical information, little is known about the spatial organization of snRNPs in the cytoplasm. We have used Larix microsporocytes as a model system to examine the organization of cellular snRNPs. In the cytoplasm of the larch microsporocytes, a cyclical detection pattern of bodies containing snRNA and Sm proteins was observed during anther meiosis. Here, we show that cyclic changes in the levels and distribution of Sm proteins are accompanied by the presence of bodies containing snRNA and Sm proteins in the cytoplasm and then an accumulation of snRNP occurs in the nucleus, leading to the formation of new Cajal bodies.
The mechanism responsible for managing the formation of Cajal bodies is coupled to the snRNP pathway. Such a conclusion is reached on the basis of the results from experiments using heterokaryons obtained by the fusion of primary human foreskin fibroblasts (DFSF1) with HeLa cells (Sleeman et al. 2001). During normal growth, DFSF1 cells are characterized by a lack of Cajal bodies. However, Cajal bodies form in the nuclei soon after fusion with HeLa cells. It was also determined that the exogenous overexpression of the SmB protein was sufficient for periodic induction of Cajal bodies in primary fibroblast cells (DFSF1).
To determine whether other snRNP factors observed in the Cajal bodies could induce de novo CB formation in larch, a U6 snRNA analysis was performed. Contrary to the other four “spliceosomal RNAs”, U6 snRNA is synthesized by RNA polymerase III (Kiss et al. 1987). In addition, U6 snRNA does not possess the trimethylguanosine cap at the 5′ end (Ro-Choi 1999). However, like the other snRNAs, its maturation and binding to other spliceosomal factors occurs in the Cajal bodies (Stanek and Neugabauer 2004, 2006). The nucleoli represent the first compartment to which U6 snRNA travels after its synthesis (Lange and Gerbi 2000). The inner nucleotides undergo methylation and pseudouridylation with the participation of snoRNA (Tycowski et al. 1998; Ganot et al. 1999). Further in vitro studies have demonstrated that after several hours, the U6 snRNA is found in the CB (Gerbi and Lange 2002), where binding to U4 and U5 snRNA takes place and where the formation of the functional tricomplex occurs (Stanek et al. 2008).
In this study, we have investigated in a natural model the hypothesis that high Sm protein expression is responsible for managing the formation of Cajal bodies. In this model, CB formation is coupled to the snRNP pathway, which has not been previously demonstrated in plants.
In our model, larch microsporocytes exhibit natural fluctuations in levels of RNA metabolism. In the larch, meiosis is characterized by a 6-month-long period and a high anther synchronization level (above 95%). In these cells, the synchronous development and “timely and fast switching on and off” of RNA and protein synthesis allows the tracking of changes that occur during snRNA synthesis and maturation under “natural conditions”.
Materials and methods
Plant material
Anthers of Larix decidua Mill. were taken from the same tree in successive G2 and meiotic prophase stages, from leptotene to late pachytene, to ensure constant experimental conditions.
Antibodies
mAb 7.13 was a gift from Prof. F. Ramaekers, Department of Pathology, University of Nijmegen, The Netherlands, which was kindly provided by Dr. JHS Scheel, University of Wageningen, The Netherlands. The human polyclonal antibody AF-ANA recognizing the larch Sm proteins (Niedojadło and Górska-Brylass 2003) was a gift from Dr. Pombo, University of Oxford. mAb Y12 was a gift from Dr. K. Neugebauer, Max Planck Institute, Dresden, Germany and was kindly provided by Dr. J Niedojadło, N. Copernicus University, Toruń, Poland. The mouse monoclonal 4G3 antibody (Acris Antibodies, Herford, Germany) recognizing the spliceosomal protein U2B″ (Habets et al. 1989; Boudonck et al. 1998) was used. The mouse anti-2,2,7 trimethyl guanosine (m3G) antibody from Calbiochem (clone K121) was used. This antibody cross-reacts with the specific 5′ cap of small nuclear RNAs (snRNAs).
Immunoprecipitation
Immunoprecipitation assays were carried out using nuclear protein extract from larch anthers. Fresh material was ground to a fine powder in liquid nitrogen. Nuclei were isolated using the CellLytic Plant Nuclei Isolation/Extraction Kit (Sigma, St. Louis, MO, USA), according to the manufacturer’s instructions.
Purified nuclei were resuspended in a buffer containing 40 mM Tris–HCl, pH 8.0, 420 mM NaCl, 1.5 mM Mg2Cl2, 0.2 mM EDTA, 0.5 mM DTT, and 0.5 mM PMSF. The nuclei were subsequently shaken for 30 min and centrifuged (800g) for 5 min, and the nuclear protein extracts were isolated. For the immunoprecipitation procedures, we used the mouse anti-SmD 7.13 antibody. Briefly, the primary antibody (anti-SmD) and 1 mM PMSF, 1% Triton X-100, and 0.5% Nonidet P-40 were added to the nuclear protein extract and incubated for 4 h at 4°C. The sample was gently mixed using an orbital shaker. The antibodies were then bound to Protein A-Sepharose beads (Sigma) and incubated for 2 h at 4°C with 60% protein A-Sepharose in TBS and NP-40. The samples were centrifuged at 16,000g for 15 min at 4°C. The pellet was then washed with 1 ml washing buffer (TBS, 1% Triton X-100, and 0.5% Nonidet P-40). This step was repeated three times. Finally, the resulting pellet was resuspended in 50 μl Laemmli buffer and used for the SDS-polyacrylamide gel electrophoresis (PAGE) and western blotting procedures. SDS-PAGE was performed using 12% acrylamide gels with 4.5% stacking gels, using the Bio-Rad Mini-Protean equipment. Antigen–antibody complexes and total protein extracts were fractionated by SDS-PAGE (12%) and transferred to PVDF membranes. The membranes were incubated overnight at 4°C with the appropriate primary antibodies, specifically, anti-SmD diluted 1:5,000 or mouse anti-TMG, diluted 1:5,000. The membranes were washed with TBS (three times), probed with an anti-mouse secondary antibody (1:1,000) for 1 h, and developed using enhanced chemiluminescence (ECL system, Amersham).
Y12 Ab specificity assay
Total proteins were isolated from larch anthers. The ground tissue was resuspended in extraction buffer (100 mM Tris–Cl, pH 7.5, 1 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail, Sigma), gently shaken, and clarified by centrifugation at 16,000g for 30 min at 4°C. The supernatant was used for SDS-PAGE and immunoblotting using Y12 Ab (diluted 1:500). The membrane was then washed with TBS (three times), probed with an anti-mouse secondary antibody (1:1,000) for 1 h, and developed using enhanced chemiluminescence (ECL system, Amersham).
Standard transmission electron microscopy
Samples were processed for electron microscopy by conventional techniques (paraformaldehyde-osmium fixation and lead citrate-uranyl acetate staining) (Niedojadło and Górska-Brylass 2003).
Isolation of meiotic protoplasts
Anthers were fixed in 4% paraformaldehyde in PBS (pH 7.2) for 6 h and crushed to obtain free meiocytes. The cells were then centrifuged and resuspended in hydrolyzing medium, consisting of 4% cellulase Onozuka R10 (Serva, Heidelberg, Germany) and 27 U/ml pectinase (Sigma) in 0.01 M citric buffer (pH 4.8) for 45 min at 37°C. The suspended protoplast matter was spread on gelatin-coated slides that were placed on dry ice and air-dried. Next, the protoplasts were rinsed with 0.1% Triton X-100 in PBS (pH 7.2) for 10 min and air-dried. These protoplasts were then used for immunodetection of Sm, m3G snRNA, and U2B″ and for fluorescence in situ hybridization of SmD1 mRNA, poly(A) RNA, U1 snRNA, U2 snRNA, and U6 snRNA.
Design of double-labeling reactions
Several double-labeling immunofluorescent-fluorescent in situ hybridization (FISH) or high-resolution immunogold in situ hybridization (HISH) reactions (Sm-U1, U2, and U6 snRNA; Sm-U2B″; BrU-poly(A) RNA; m3G snRNA-U2 snRNA) were performed as described below. In all experiments, immunocytochemistry always preceded in situ hybridization because immunofluorescent signals were very weak when ISH was applied first. In the double-labeling immunofluorescence method (m3G snRNA-Sm or U2B″-Sm), both primary and secondary antibodies were applied simultaneously in the incubation medium.
Immunodetection of Sm proteins, U2B″ and m3G snRNA
The immunofluorescent and immunogold SmD localization experiments were performed using the mouse 7.13 antibody (1:10) (Testillano et al. 1993). For Sm, we used a human αSm autoimmune serum against Sm that recognizes the plant Sm protein (1:200) (Pálfi et al. 1989) according to a schedule established by Wróbel and Smoliński (2003). The mouse monoclonal antibody 4G3 (1:10) (Organon Teknika) recognizing the spliceosomal protein U2B″ was used (Habets et al. 1989; Boudonck et al. 1998). For the m3G cap, we used an anti-m3G antibody (Calbiochem, Bad Soden, Germany) according to the protocol provided by Zienkiewicz et al. (2006). Sm proteins were also detected by incubation with primary mouse anti-Sm Y12 according to the method of Zienkiewicz et al. (2008) and/or with primary AF-ANA human anti-Sm antibody in 0.2% acetylated BSA in PBS (1:100) in a humidified chamber at 8°C overnight and an anti-human antibody Cy3 (Sigma) secondary antibody according to Kołowerzo et al. (2009).
Quantitative measurements
An image analysis was performed on larch microsporocyte protoplasts after immunofluorescence staining using mAb Y12, with each reaction step performed using consistent values of temperature, incubation times, and concentrations of primary and Cy3 secondary antibodies. Thirty-five cells in each stage from three different preparations were analyzed. The results were registered using a Nikon C1 confocal microscope with a He–neon laser emitting light with a wavelength of 543 nm (green excitation and yellow fluorescence). The three-dimensional optical sections were acquired with a 0.2-μm step interval. For all antigens and developmental stages, the obtained data were corrected for background autofluorescence determined by negative control signal intensities. For the image processing and analysis, the EZ Viewer software package (Nikon Europe BV, Badhoevedorp, The Netherlands) was used. The analysis was performed using Lucia General software (Laboratory Imaging, Prague, Czech Republic) compatible with a Nikon PCM 2000 confocal microscope and Nikon C1 confocal microscope. To test differences among multiple samples (groups, i.e., Sm level in different stages), a Kruskal–Wallis ANOVA test was used.
FISH in double-labeling reactions
For hybridization, the probe was resuspended in hybridization buffer (30% v/v formamide, 4× SSC, 5× Denhardt’s buffer, 1 mM EDTA, and 50 mM phosphate buffer) at a concentration of 50 pmol/ml. Hybridization was performed overnight at 37°C. The following DNA oligonucleotides were used: antisense U2 snRNA—5′ Alexa 594 ATATTAAACTGATAAGAACAGATACTACACTTG 3′, sense U2 snRNA—5′ Alexa 594 CAAGTGTAGTATCTGTTCTTATCAGTTTAATAT (IBB PAN, Warsaw, Poland), antisense U1 snRNA—5′ Alexa 594 ACAGCAACAAATTATGATGTAGGTTC 3′, sense U1 snRNA—5′ Alexa 594 GAACCTACATCATAATTTGTTGCTGT 3′ (IBB PAN, Warsaw, Poland), antisense U6 snRNA—5′ Alexa 594 CTAATCTTCTCTGTATCGTTCCAATTTTA 3′, sense U6 snRNA—5′ Alexa 594 TAAAATTGGAACGATACAGAGAAGATTAG 3′ (IBB PAN, Warsaw, Poland)
ISH immunogold monolabeling of U2 snRNA and double labeling with Sm proteins
After fixation in 4% PFA in PBS overnight, anthers were rinsed in PBS and dehydrated and mounted in LRGold resin (Sigma), according to the Majewska-Sawka and Rodriguez-Garcia (1996) schedule. Ultrathin sections were collected on Formvar-coated nickel or gold grids. Ultrathin sections were pretreated with pre-hybridization buffer (hybridization buffer without probe) for 1 h at room temperature and then incubated with the antisense U2 snRNA probe (5′ ATATTAAACTGATAAGAACAGATACTACACTTG 3′) in hybridization buffer (the probe was labeled at the 3′ end with digoxigenin nucleotides by terminal deoxynucleotidyl transferase) (Roche). Hybridization was performed in a sealed, humidified chamber for 20 h at room temperature. Post-hybridization washing was performed as described by Smoliński et al. (2007). Non-specific antigens were blocked with PBS buffer containing 0.05% acetylated BSA for 0.5 h. DNA–RNA hybrids were localized by incubation with sheep anti-digoxigenin antibody coupled to 10-nm-diameter colloidal gold particles (Orion Diagnostica, Espoo, Finland) diluted 1:30 in PBS containing 0.02% acetylated BSA for 1 h at room temperature. Grids were then rinsed and contrasted according to Smoliński et al. (2007).
In the double-labeling reactions, immunogold localization of Sm proteins was performed prior to ISH immunogold labeling, using the primary AF-ANA human anti-Sm protein B/B′, D1 antibody in 0.2% acetylated BSA in PBS (1:100) at 8°C in a humidified chamber overnight. Samples were incubated with secondary anti-human antibody coupled to 15-nm-diameter colloidal gold particles (BioCell, Cardiff, UK) and rinsed and contrasted (1% phosphotungstic acid and 2.5% uranyl acetate).
Microscopy
The results were registered with Nikon C1 and Nikon PCM 2000 confocal microscopes using an argon–ion laser emitting light with a wavelength of 488 nm (blue excitation and green fluorescence), a He–neon laser emitting light with a wavelength of 543 nm (green excitation and red fluorescence), and a He–neon laser emitting light with a wavelength of 594 nm (red fluorescence). A mid pinhole, long exposure time (75 μs) and a 60× (numerical aperture, 1.4) Plan Apochromat DIC H oil immersion lens was used. Pairs of images were collected simultaneously in the green (Alexa 488 or FITC fluorescence) and red (Cy3 or Alexa 594) channels. To minimize bleed-through between the fluorescence channels, we applied low laser power (3–10% of maximum power) and single-channel collection. For the bleed-through analysis and control experiments, Lucia G software was used (Laboratory Imaging, Prague, Czech Republic). For DAPI staining, an inverted Nikon Eclipse TE 2000 fluorescence microscope equipped with a mercury lamp, a UV-2EC UV narrow-band filter, and a DXM 1200 FX digital camera was used. Nikon NIS-elements deconvolution software were then used.
The material was examined at the ultrastructural level, and photomicrographs were taken, using a Jeol 1010 (Japan) electron microscope at 80 kV.
Control reactions
For in situ hybridizations (high resolution) and immunofluorescence methods, control treatments consisting of incubations without the primary antibody were performed. For the in situ hybridizations—high-resolution and fluorescence analyses, sense-labeled probes and ribonuclease-treated samples were used as additional controls. All control reactions produced negative results, or the results of the control reactions were very low compared with the standard reactions.
Results
Determination of Sm and TMG snRNA antiserum reactivity
We used the mouse mAb 7.13, which recognizes the SmD protein of the Ul, U2, U4, U5, and U6 snRNPs in plants (Billings et al. 1982, 1985; Pálfi et al. 1989; Testillano et al. 1993; Straatman and Schel 2001). Immunoprecipitation and western blot analyses demonstrated that the mouse monoclonal anti-SmD antibody recognized a band with a molecular mass of about 55 kDa (Fig. S1). As a control, we used a primary monoclonal anti-SmD antibody without sample adding and we tested its direct cross-reactivity with the secondary Ab following the IP procedure.
Three bands were visible after immunoprecipitation with the anti-SmD antibody and immunoblotting with the SmD and anti-TMG cap Abs. Two prominent bands (60 and 28 kDa) represent the cross-reactivity after immunoblotting with primary anti-SmD and secondary antibody. These two prominent bands represent the heavy (upper band) and light (lower band) chains of the antibody, respectively. The weaker band cross-reacts with the primary anti-Sm Ab and the secondary antibody and represents the immunoprecipitated protein.
Similar results were obtained with an anti-TMG snRNA antibody, which recognized the same complex with a molecular mass of 55 kDa. This result indicated that the 55-kDa moiety was a snRNP complex containing SmD proteins and snRNA.
In the case of the anti-TMG cap Ab, two prominent bands also represent the heavy (upper band) and light (lower band) chains of the SmD antibody, which cross-react with secondary antibody. However, the weaker band cross-reacts with the primary anti-TMG Ab and represents the immunoprecipitated Sm proteins complexed with snRNA containing the TMG cap structure.
Moreover, because the two primary antibodies (SmD and anti-TMG) were obtained from mouse, the cross-reactivity observed in lane 3 also represents the heavy and light chains after probing with secondary (anti-mouse) Ab labeled with horseradish peroxidase.
The two bands are more prominent because the concentration of the SmD antibody used for immunoprecipitation was much higher than the cellular concentration of immunoprecipitated antigen in the larch anthers.
A western immunoblot analysis was performed to examine the specificity of the mouse Y12 antibody, in Larix decidua which recognizes Sm B′/B and D1 proteins in plants (Hirakata et al. 1993; Echeverría et al. 2007). As shown in Fig. S2, the mouse Y12 antibody recognized proteins of about 30 and 14 kDa, which appear to represent the methylated SmB\B′ and SmD1 proteins, respectively.
Localization of splicing factors during cyclic fluctuations in Sm protein quantities
In the analyzed stages, i.e., from G2 through pachytene, we could distinguish three cycles in which the levels and distribution of Sm proteins differed. Four A–D stages were distinguished in all three of the cycles. We named them A1–D1 in the first (G2) cycle, A2–D2 in the second (leptotene) cycle, and A3–D3 in the third (zygotene) cycle. The characteristic changes in the distribution of Sm proteins during the four stages of each cycle were demonstrated with the example of the first cycle of the G2 phase of premeiotic interphase. Two simultaneous analyses of the colocalization Sm proteins with m3G snRNA and with U6 snRNA were performed. Similar changes in snRNP distribution were observed in the two other prophase cycles (leptotene and zygotene one).
In the first stage (A1), the levels of Sm proteins were very low. In the cytoplasm and nucleoplasm, Sm proteins were present in a dispersed form (Figs. 1a, S3a). In some cases, small quantities of Sm proteins, in the form of a dispersed signal, were also observed in the nucleolus. The levels of m3G snRNA were also low at this stage (Fig. S3b), and the signal was dispersed in the nucleoplasm (Fig. S3b). The levels of U6 snRNA were also low, and U6 snRNA was mainly detected in the nucleoli (Fig. 1b).
Localization of Sm proteins (mAb Y12) and U6 snRNA during premeiotic interphase. Initially, very low levels of the Sm protein were observed (a). U6 snRNA was clearly visible only in the nucleoli (b, c). In the subsequent stage, cytoplasmic bodies containing Sm proteins appeared (e). In this period, an increase in the levels of these proteins was observed in the nucleus (e, g). The amount of U6 snRNA increased considerably in the nucleoli, but a significant amount was also detected in a dispersed form in the nucleus (f). In the next stage, when cytoplasmic bodies containing Sm proteins could still be observed in the cytoplasm, high Sm protein levels were observed in the nucleoplasm and in the nuclear bodies (i, k). Colocalization of U6 in the nuclear bodies was not observed. The levels of U6 in the nucleolus were still much higher than in the nucleoplasm (j). In the last stage, only very small, single Sm bodies were visible in the cytoplasm (arrow head). In the nucleus, strong Sm protein (m) and U6 snRNA (n) signals could be observed in the nuclear bodies (arrows), whereas the signal was uniform and dispersed in the nucleoplasm (o). The corresponding DAPI images were collected using widefield fluorescence and deconvolution software (d, h, l, p). Bars 10 μm
In the next stage (B1), a significant increase in the level of Sm proteins was observed, and fine Sm bodies began to appear in the cytoplasm, some of which contained not only Sm proteins (Figs. 1e, S3e) but also m3G snRNA (Fig. S3f). The oval clusters of Sm proteins occurring in the cytoplasm of larch microsporocytes were called cytoplasmic snRNP bodies (CsBs). Many small, fine clusters of Sm proteins or dispersed Sm signals, which did not contain m3G snRNA, formed in the cytoplasm as well (Fig. S3g). Accumulation of Sm proteins (Fig. S3e) and m3G snRNA (Fig. S3f) was observed at the border between the nucleus and the cytoplasm or at the periphery of the cell nucleus (Fig. 1e) during this phase. A significant increase in levels of U6 snRNA was also observed in the nucleoli. In the nucleolus, the U6 snRNA exhibited a dispersed pattern (Fig. 1f).
In the third stage (C1), significant quantities of Sm and m3G snRNA were still observed in the cytoplasm in the form of large, CsBs (Figs. 1i, S3i–k), and Sm proteins reached their highest level in the nucleus (Fig. 1i). Sm proteins were present in the whole nucleus. Nuclear bodies were present in high numbers at stage C (Fig. 1i). The levels of U6 in the nucleolus were still significantly higher than in the nucleoplasm (Fig. 1j). We did not observe any accumulation of U6 snRNA in the nuclear bodies containing Sm proteins (Fig. 1j, k).
In the last stage (D1), there was a significant decrease in the levels of Sm proteins and m3G snRNA in the cytoplasm (Figs. 1m, S3m–o). In the cytoplasm, only single spots containing both Sm proteins and m3G snRNA were visible (Fig. S3m–o). In the nucleus, besides the evenly dispersed signal in the nucleoplasm, the Sm proteins accumulated mainly in numerous nuclear bodies (Figs. 1m, S3g). The maximal number and size of the nuclear bodies was reached in stage D1. U6 snRNA was present at similar levels in the nucleolus and the nucleoplasm; however, the highest levels were observed in the nuclear bodies (Fig. 1n). At this stage, a high level of colocalization of Sm proteins with U6 snRNA occurred in the nuclear bodies and in the nucleoplasm (Fig. 1o).
Cytoplasmic distribution of Sm proteins in larch microsporocytes
An analysis using the double immunofluorescence/FISH labeling confirmed that CsBs could contain individual types of snRNA. The colocalization of Sm proteins with U2 snRNA (Fig. 2a–d) and with U1 snRNA (Fig. 2e–h), demonstrated that some of CsB bodies containing Sm proteins also contained detectable levels of individual analyzed snRNAs. In addition to CsB localization, U1 and U2 snRNA were present as dispersed signals in the cytoplasm (Fig. 2b, f).
Colocalization of U2 snRNA (a–d) and U1 snRNA (e–h) with Sm proteins (mAb 7.13) during premeiotic interphase (immunofluorescece/FISH method). Many of cytoplasmic structures contained Sm proteins; some CsBs posses only a small amount of snRNA or are devoid of snRNA (g). The corresponding DAPI images were collected using widefield fluorescence and deconvolution software (d, h) Bars 10 μm
The distribution of Sm proteins exhibited a characteristic pattern in the cytoplasm of the microsporocytes. During periods when Sm protein levels in the microsporocyte were high (stages B and C), Sm proteins were frequently observed in the form of CsBs. Such bodies were detected during late G2 phase (Fig. 3a), late leptotene (Fig. 3e), and middle zygotene (Fig. 3i). An analysis using the double immunofluorescence labeling method demonstrated that many Sm protein-containing cytoplasmic bodies also contained small nuclear RNA. On the basis of the colocalization of Sm proteins with m3G snRNA, we determined that mature snRNAs can be found in most cytoplasmic Sm clusters (Fig. 3). In the cytoplasm, in addition to large CsB complexes, snRNA was also detected in the form of small grains or spots, but a dispersed localization of m3G snRNA was not observed (Fig. 3b, f, j).
Immunolocalization of Sm (AF-ANA) proteins and m3G snRNA in the G2 (a–d), leptotene (e–h) and zygotene (i–l) phases using the double-label immunofluorescence method. Large spherical cytoplasmic bodies (arrows) containing both Sm proteins and snRNA can be observed. During this period, numerous smaller clusters of Sm proteins were also present in the cytoplasm (arrowheads), but not always together with snRNA. The corresponding DAPI images were collected using widefield fluorescence and deconvolution software (d, h, l). Bars 10 μm
An electron microscopy analysis allowed us to understand the ultrastructural characteristics of the CsBs that were observed in larch microsporocytes. Immunogold localization with the Y12 anti-Sm antibodies revealed that these oval bodies are the sites of the selective accumulation of Sm proteins (Fig. 4a). Therefore, these oval bodies correspond to the CsBs that were observed by confocal microscopy. In addition to the large bodies, the gold particles labeling the Sm proteins were also found in a dispersed form and in sparse clusters in the cytoplasm (Fig. 4a). Cytoplasmic bodies were also strongly labeled with the anti-m3G snRNA antibodies (Fig. 4b). These bodies were the main area of m3G snRNA accumulation in the microsporocyte cytoplasm. No scattered m3G snRNA signals were observed, whereas scattered signals were visible in the case of Sm proteins (Fig. 4a, b).
Ultrastructural identification of cytoplasmic snRNP bodies. a Immunogold labeling of Sm proteins (mAb Y12). Two cytoplasmic bodies (CsB) in close contact (arrows) with a large accumulation of Sm proteins were observed occasionally. In the cytoplasm, the signal was more scattered than in the CsB. b m3G snRNA. snRNA is preferentially localized in the CsB and in several smaller clusters in the cytoplasm (dashed line). c Colocalization of U2 snRNA (10 nm) and Sm proteins (15 nm) immunogold/high resolution ISH method. U2 and Sm can be localized in various zones of the bodies. In the nucleoplasm, the majority of Sm protein clusters also contained the U2 snRNA (arrowheads). Sometimes, the Sm proteins (asterisk) were observed alone. Bars 0.5 μm
The presence of snRNA in the analyzed Sm-containing CsBs was confirmed through the use of a colocalization immunogold/ISH immunogold technique for Sm (Y12) and U2 snRNA (Fig. 4c). Furthermore, the cytoplasmic colocalization of Sm proteins and U2 snRNA was also observed in fine clusters (Fig. 4c).
Nuclear localization of Sm proteins in Cajal bodies
In microsporocyte nuclei, the distribution of Sm proteins often had a dispersed character (phase C, Figs. 1i, 3i), or an accumulation of Sm-snRNP occurred in the nucleus (phase D, Figs. 1m, S3m). Oval nuclear bodies (0.5–3 μm diameter) containing the Sm and U2B″ proteins and other splicing factors were often observed (Fig. 5a, b). Nuclear bodies with high concentrations of Sm proteins and snRNA were detected during stages from the G2 phase of premeiotic interphase through the prophase of the first meiotic division.
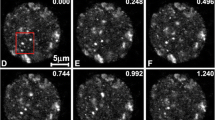
The Cajal bodies of larch microsporocytes. In the nuclei, spherical nuclear bodies were present, exhibiting a regular shape and characterized by labeling of Sm (a) and U2B proteins (b) at much higher levels than in the surrounding nucleoplasm (a, b). Bar 10 μm. c Standard electron microscopy (EM) technique. The Cajal bodies (CB) are made up of coiled fibrils of 15–25 nm in diameter, with nearby visible clusters of interchromatin granules (IG). Immunolocalization of SmD proteins (mAb Y12) (d). A cluster of intensely labeled Sm proteins was detected in the Cajal body close to the nuclear envelope and IG (d). Colocalization of U2 snRNA (10 nm gold particles) and Sm proteins (15 nm gold particles) (αSm) at the ultrastructural level (e) immunogold/high-resolution ISH method. In the Cajal bodies, labeling and colocalization of Sm proteins with U2 snRNA was very high (e). The localization of m3G snRNA (15 nm gold particles) and U2 snRNA (10 nm) labeling in the Cajal bodies is much stronger than in the surrounding nucleoplasm (f) immunogold/high-resolution ISH method. Bars 0.5 μm
Nuclear bodies consisting of coiled fibrils, resembling a coil of twisted thread with dense coiled fibrils (with a diameter of 15–25 nm), were observed in larch microsporocytes (Fig. 5c). This substructure is a characteristic trait of the Cajal bodies found in plants and animals. The application of the immunogold technique at the ultrastructural level demonstrated that Cajal bodies were the place of Sm protein accumulation (Fig. 5d). Therefore, Cajal bodies corresponded to the oval clusters of signal that were observed by confocal microscopy. CBs were very often localized to various parts of the nucleus close to the nuclear envelope or near clusters of interchromatin granules (IGs) (Fig. 5d). The presence of U2 snRNA, m3G snRNA, and Sm proteins was demonstrated at the ultrastructural level in Cajal bodies using a double-labeling immunogold/ISH immunogold method (Fig. 5e, f). There was an almost equal distribution of Sm proteins and snRNA (Fig. 5e, f). The levels of Sm proteins and snRNA were many times higher in Cajal bodies than in the nucleoplasm, a finding that is especially apparent at the ultrastructural level (Fig. 5d–f). We found that Sm proteins and U2 snRNA were distributed preferentially in the CB (412.6 ± 96 and 149.5 ± 8.1 gold grains/μm2, respectively), at significantly higher levels (p < 0.05) when compared with nucleoplasm areas (19.5 ± 7.2 grains/μm2 and 6.7 ± 0.9 gold grains/μm2) during the G2 stage. We obtained similar results from a quantitative evaluation of Sm proteins and U2 snRNA distribution during the leptotene and zygotene cycles.
Levels of Sm proteins during the G2 phase of premeiotic interphase and early prophase during the first meiotic division
In this study, we performed a quantitative analysis of the emitted fluorescence from the snRNPs complexes. Immunofluorescence using anti-Sm Y12-Cy3 was employed to demonstrate that the levels of Sm proteins undergo significant fluctuations during the premeiotic G2 phase of interphase through the later pachytene in larch microsporocytes (Fig. 6). The stages of premeiotic interphase and meiosis were recognized on the basis of chromatin structure (DAPI staining) and measurements of nuclear volume and whole cell volume (during the studied period, the whole nuclear volume increased by over fourfold and the whole cell volume increased by more than fivefold).
The levels of Sm proteins (Y12) in the period from the G2 phase of the premeiotic interphase to pachytene in larch microsporocytes (parallel results were obtained using mAb 7.13 and AF-ANA). Three similar cycles of the amount and distribution of Sm proteins were highlighted, and four stages within each cycle were distinguished
High levels of Sm proteins were observed during three phases of microsporocyte development. The first of these phases occurred in interphase, at the end of the G2 phase. The other two phases were observed during late leptotene and middle zygotene (Fig. 6).
Discussion
Studies have revealed that during anther meiosis in the larch, spherical structures, which are not separated by a membrane, periodically appear in the cytoplasm. The absence of a membrane surrounding these cytoplasmic bodies suggests that a special mechanism must exist that facilitates their formation and protects them from the cellular autolytic system. Our studies have demonstrated that cytoplasmic bodies, which contain Sm proteins and snRNA, exist in the cytoplasm of larch meiocytes. These structures were named “cytoplasmic snRNP bodies” (CsBs). Until now, there have been no reports on similar phenomena in higher plants.
In animal germ cells, cytoplasmic granules that possess similar structural properties are called nuage (Chuma et al. 2003), P bodies (Eulalio et al. 2007) or P-granules (Schisa et al. 2001), and some of them may contain Sm proteins (Biliński et al. 2004).
In Xenopus oocytes, nuage contain not only the spliceosomal Sm proteins but also Xcat2 mRNA. Other components of Cajal bodies or the splicing machinery, such as coilin, SMN protein, and snRNAs were absent from the nuage (Biliński et al. 2004). The authors suggested that Xenopus Sm proteins have adapted to a role independent of pre-mRNA splicing and that instead of binding to their traditional spliceosomal partner, such as snRNA, they bind mRNAs that are components of the germinal granules (i.e. Xcat2 mRNA) and facilitate the transport of these mRNAs from the nucleus to the nuage, which are precursors of germinal granules.
Studies in C. elegans show that Sm proteins are components of P-granules that are required for germ-granule (P-granules) localization (Barbee et al. 2002; Barbee and Evans 2006). Non-spliceosomal roles for Sm proteins were also postulated during the maturation of spermatocytes and for germ-cell specification in Drosophila (Gonsalvez et al. 2006). Finally, Gonsalvez et al. (2010) have demonstrated that SmB and SmD3 are specific components of the oskar messenger ribonucleoprotein (mRNP) in Drosophila.
In the nuclei of Drosophila oocytes Liu and Gall (2007) have observed that snRNA, Sm, and SMN proteins accumulate occasionally in structures that the authors called U bodies, which are located near P bodies. These structures could correspond to CsBs in the larch. The authors suggest that U bodies may be the sites for some steps of snRNP assembly or that they may serve primarily for the storage of snRNPs after assembly but before import into the nucleus. Further studies by the same authors (Lee et al. 2009) have demonstrated a relationship between SMN proteins that participate in snRNP complex maturation and the integrity of both U bodies, which contain this type of snRNP, and P bodies, which participate in the regulation and degradation of other types of RNAs in the cytoplasm.
Cytoplasmic bodies that exist in the cytoplasm of larch meiocytes contain snRNA in addition to Sm proteins. The presence of Sm–snRNP in larch CsB could be in agreement with the sequence of molecular events in the snRNP cellular cycle. In the cytoplasm, a cortex molecule comprised of Sm proteins attaches to snRNA, the cap is hypermethylated, and the 3′ end matures (Will and Luhrmann 2001; Massenet et al. 2002).
Cytoplasmic snRNP bodies could serve as potential platforms for the formation of this complex in larch meiocytes. The colocalization of Sm proteins and snRNA in the larch cytoplasmic bodies supports such a hypothesis. In addition to bodies that contained both molecules, foci were also found that contained only Sm proteins. These small foci could represent compartments where the Sm cortex molecule is stored, or they could be a possible place for intensive synthesis of the Sm cortex molecule. The localization of SmD1 mRNA proved its local accumulation in the cytoplasm, which could correspond to the translation site of Sm proteins (data not shown). Using the hybridization method, we demonstrated that the U1 and U2 snRNAs exist in a dispersed form and in CsB clusters, where it almost always colocalizes with the Sm proteins. Slightly different patterns of localization were observed for snRNA detected by antibodies against the m3G cap. In this method, trimethylated snRNAs were observed in the cytoplasm. In the cytoplasm, using the colocalization method, m3G snRNA was found mainly in CsB clusters and always with Sm proteins. Almost no m3G snRNA was observed in the cytoplasm in a dispersed form or without colocalized Sm proteins. These observations indicate that after export to the cytoplasm snRNAs can travel in a dispersed form and then bind to small clusters of Sm proteins and form CsBs. After dimethylation, which presumably takes place in the CsB, snRNP complexes can move toward the nucleus.
Increase of Sm proteins and snRNA content and the formation of CsBs in all three cycles always preceded the appearance of Cajal bodies. Such a positive correlation indicates that the occurrence of both phenomena is not a random coincidence. To determine whether other snRNP factors observed in the Cajal bodies could induce de novo CB formation in larch, a U6 snRNA analysis was performed. Our observations have shown that in the early stages of the analyzed cycles, the U6 snRNA was detected in the nucleoli at the same time that the Sm-containing CsBs appeared. In the subsequent stages of the cycle, when the first Sm-containing CBs and m3G snRNA began to appear in the nucleoplasm, the U6 snRNA is localized, in addition to in the nucleolus, in a dispersed form in the nucleoplasm, but not in the CBs. The accumulation of U6 snRNA in the CBs was observed only in the later part of the cycle, when numerous and large CBs had already appeared in the nuclei. Contrary to the other spliceosomal RNAs, U6 snRNA is not necessary in the initial step of Cajal body formation in larch.
The formation of CsBs always preceded the appearance of Cajal bodies. Such a positive correlation indicates that the occurrence of both phenomena is not a random coincidence. High expression of Sm proteins, demonstrated by the appearance of foci containing this protein in the cytoplasm, induces the assembly of Sm–snRNP complexes in the cytoplasmic snRNP complex, which is where snRNA dimethylation likely occurs. In the next stage most likely cytoplasmic bodies transport to the nucleus membrane occur, which is evidenced by the accumulation of Sm proteins and CsB at the nucleus–cytoplasm border. A similar “bodies like” pathway of transport is also pointed out by the ultrastructural images of Xenopus oocytes containing nuage (Biliński et al. 2004). In larch, this type of transport is also supported by the lack of a dispersed localization of m3G snRNA in the cytoplasm. If the cytoplasmic/nuclear Sm–RNPs stay in a dynamic balance, a high level of Sm protein synthesis in the cytoplasm may cause an elevated accumulation of Sm–snRNP in the nucleus. A morphological manifestation of this process seems to be observed as the accumulation of diffusionally dispersed Sm–snRNP in the sub-membrane space to the meiocytic nuclei. Exceeding the Sm–snRNP concentration over the threshold can trigger the interaction of these complexes with coilin homologues, whose presence has been confirmed in plants (Collier et al. 2006; Koroleva et al. 2009).
The interaction between the snRNP cycle and Cajal body formation has also been observed in the earlier studies in HeLa cells, where blockade of hTGS1, SMN, and PHAX protein expression was achieved by RNAi (Lemm et al. 2006). The knockdown of these three proteins disturbed the maturation of UsnRNPs before its re-importation into the nucleus and caused a decrease in Cajal body formation. A similar decrease in the quantity of CBs has also been observed in HeLa cells exposed to leptomycin B (Sleeman et al. 2001). Leptomycin B is a compound that blocks snRNP export from the nucleus to the cytoplasm as well as its re-importation into the nucleus. This decrease in snRNP levels in the nucleus has been attributed to the inhibition of CB formation or to CB disorganization.
Lemm et al. (2006) have suggested the following model for the formation of Cajal bodies: (1) during the first stage, snRNP core molecules, which consist of Sm proteins and snRNA, interact with coilin in the nucleoplasm. (2) In the next stage, the self-oligomerization of coilin induces the joining of various factors that are related to snRNP splicing, and (3) in the final stage of assembly, other domains such as snoRNP are also built, which finally results in the formation of CBs.
Recently, Strzelecka et al. (2010a) showed that coilin and snRNPs are concentrated in numerous CBs in zebrafish embryos before, during, and after activation of the zygotic genome. They suggested that CB number is regulated during development to respond to the demands of gene expression in a rapidly growing embryo. Following zygotic genome activation, snRNP biogenesis was required for CB assembly and maintenance, suggesting a self-assembly process that determines CB numbers in embryos.
In our embryological model, larch microsporocytes also exhibit natural fluctuations in levels of mRNA and rRNA metabolism (Smoliński et al. 2007; Smoliński and Kołowerzo 2011). In the larch, the synchronous development and “timely and fast switching on and off” of RNA and protein synthesis allows the tracking of changes that occur during snRNA synthesis and maturation snRNP cycle, and Cajal body formation has also been observed. Further study (Strzelecka et al. 2010b) showed that also coilin is required during vertebrate embryogenesis to maintain the supply of assembled spliceosomal snRNPs sufficient for expression of zygotic transcripts. Depletion of coilin in zebrafish embryos leads to CB dispersal, deficits in snRNP biogenesis and expression of spliced mRNA. snRNAs were necessary but not sufficient for rescue, showing that only assembled snRNPs can bypass the requirement for coilin. Thus, coilin’s essential function in embryos is to promote macromolecular assembly of snRNPs, likely by concentrating snRNP components in CBs to overcome rate-limiting assembly steps (Strzelecka et al. 2010b).
It seems that our results agree with the recently postulated hypothesis about the self-organization of compartments such as Cajal bodies, which is due to ability to selfdimerize such molecules like coilin, Sm proteins, and SMN (Misteli 2007). Kaiser et al. (2008) have artificially induced the de novo formation of Cajal bodies on chromatin after the immobilization of structural and functional components of the CB, such as coilin, SmD1, SmE and SmG proteins, and other factors related to snRNA and snoRNA. The authors have suggested that Cajal bodies originate as self-organizing structures, which are formed as local concentrations of macromolecules, but not through a “hierarchical assembly pathway”. Finally, tethering non-CB components to the lac operator array failed to nucleate CB formation, whereas tethering of PML body components resulted in formation of de novo PML bodies (Kaiser et al. 2008). Taken together, these data strongly support a stochastic assembly model and argue against an ordered or hierarchical nuclear body assembly pathway (Matera et al. 2009). Currently, it has been suggested that other nuclear compartments related to snRNA metabolism function in a similar way to CBs. Speckles, which represent another dynamic nuclear compartment, have their own aggregation-diffusion model of organization (Carrero et al. 2005). It cannot be excluded that also cytoplasmatic compartments related to snRNA metabolism as CsBs function in a similar way to nuclear compartments.
References
Acevedo R, Samaniego R, Moreno Díaz de la Espina S (2002) Coiled bodies in nuclei from plant cells evolving from dormancy to proliferation. Chromosoma 110:559–569
Barbee SA, Evans TC (2006) The Sm proteins regulate germ cell specification during early C. elegans embryogenesis. Dev Biol 291:132–143
Barbee SA, Lublin AL, Evans TC (2002) A novel function for the Sm proteins in germ granulelocalization during C. elegans embryogenesis. Curr Biol 12:1502–1506
Batalova FM, Stepanova IS, Skovorodkin IN, Bogolyubov DS, Parfenov VN (2005) Identification and dynamics of Cajal bodies in relation to karyosphere formation in scorpionfly oocytes. Chromosoma 113:428–439
Biliński SM, Jaglarz MK, Szymanska B, Etkin LD, Kloc M (2004) Sm proteins, the constituents of the spliceosome, are components of nuage and mitochondrial cement in Xenopus oocytes. Exp Cell Res 299:171–178
Billings PB, Allen RW, Jensen FC, Hoch SO (1982) Anti-RNP monoclonal antibodies derived from a mouse strain with lupus-like autoimmunity. J Immunol 128:1176–1180
Billings PB, Barton JR, Hoch SO (1985) A murine monoclonal antibody recognizes the polypeptide of the Sm small nuclear ribonucleoprotein complex. J Immunol 135:428–432
Bogolyubov D, Parfenov V (2001) Immunogold localization of RNA polymerase II and pre-mRNA splicing factors in Tenebrio molitor oocyte nuclei with special emphasis on karyosphere development. Tissue Cell 33:549–561
Boudonck K, Dolan L, Shaw P (1998) Coiled body numbers in the Arabidopsis root epidermis are regulated by cell type, developmental stage and cell cycle parameters. J Cell Sci 111:3687–3694
Boudonck K, Dolan L, Shaw PJ (1999) The movement of coiled bodies visualized in living plant cells by the green fluorescent protein. Mol Biol Cell 10:2297–2307
Cajal SR (1903) Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab Lab Invest Biol 2:129–221
Carrero G, Hendzel MJ, de Vries G (2005) Modelling the compartmentalization of splicing factors. J Theor Biol 239:298–312
Chouinard LA (1975) A light and electron-microscope study of the oocyte nucleus during development of the antral follicle in the prepubertal mouse. J Cell Sci 17:589–615
Chuma S, Hiyoshi M, Yamamoto A, Hosokawa M, Takamune K, Nakatsuji N (2003) Mouse Tudor Repeat-1 (MTR-1) is a novel component of chromatoid bodies/nuages in male germ cells and forms a complex with snRNPs. Mech Dev 120:979–990
Cioce M, Lamond AI (2005) Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol 21:105–131
Collier S, Pendle A, Boudonck K, van Rij T, Dolan L, Shaw P (2006) A distant coilin homologue is required for the formation of cajal bodies in Arabidopsis. Mol Biol Cell 17:2942–2951
Dickmanns A, Ficner R (2005) Role of the 5′-cap in the biogenesis of spliceosomal snRNPs. Top Curr Genet 12:179–204
Echeverría O, Vázquez-Nin G, Juárez-Chavero S, Moreno Díaz de la Espina S (2007) Firing of transcription and compartmentalization of splicing factors in tomato radicle nuclei during germination. Biol Cell 99:519–530
Eulalio A, Behm-Ansmant I, Izaurralde E (2007) P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8:9–22
Ferreira J, Carmo-Fonseca M (1995) The biogenesis of the coiled body during early mouse development. Development 121:601–612
Ferreira J, Carmo-Fonseca M (1996) Nuclear morphogenesis and the onset of transcriptional activity in early hamster embryos. Chromosoma 105:1–11
Ferreira JA, Carmo-Fonseca M, Lamond AI (1994) Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J Cell Biol 126:11–23
Frey MR, Matera AG (1995) Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci USA 92:5915–5919
Ganot P, Jady BE, Bortolin ML, Darzacq X, Kiss T (1999) Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol Cell Biol 19:6906–6917
Gerbi SA, Lange TS (2002) All small nuclear RNAs (snRNAs) of the [U4/U6.U5] Tri-snRNP localize to nucleoli; identification of the nucleolar localization element of U6 snRNA. Mol Biol Cell 13:3123–3137
Gonsalvez GB, Rajendra TK, Tian L, Matera AG (2006) The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr Biol 16:1077–1089
Gonsalvez GB, Rajendra TK, Wen Y, Praveen K, Matera AG (2010) Sm proteins specify germ cell fate by facilitating oskar mRNA localization. Development 137:2341–2351
Górska-Brylass A, Wróbel B (1978) An electron microscope study of extranucleolar ribonucleoprotein bodies in the microspore of Larix europea L. Soc bot Fr Act bot 1–2:23–25
Habets WJ, Hoet MH, Sillekens PT, De Rooij DJ, Van de Putte LB, Van Venrooij WJ (1989) Detection of autoantibodies in a quantitative immunoassay using recombinant ribonucleoprotein antigens. Clin Exp Immunol 76:172–177
Hirakata M, Craft J, Hardin JA (1993) Autoantigenic epitopes of the B and D polypeptides of the U1 snRNP. Analysis of domains recognized by the Y12 monoclonal anti-Sm antibody and by patient sera. J Immunol 150:3592–3601
Huang S, Deerinck TJ, Ellisman MH, Spector DL (1994) In vivo analysis of the stability and transport of nuclear Poly(A) + RNA. J Cell Biol 4:877–899
Kaiser TE, Intine RV, Dundr M (2008) De novo formation of a subnuclear body. Science 322:1713–1717
Kiss T, Antal M, Solymosy F (1987) Plant small nuclear RNAs. II. U6 RNA and a 4.5SI-like RNA are present in plant nuclei. Nucleic Acid Res 15:543–560
Kołowerzo A, Smoliński DJ, Bednarska E (2009) Poly(A) RNA a new component of Cajal bodies. Protoplasma 236:13–19
Koroleva OA, Calder G, Pendle AF, Kim SH, Lewandowska D, Simpson CG, Jones IM, Brown JW, Shaw PJ (2009) Dynamic behavior of Arabidopsis eIF4A-III, putative core protein of exon junction complex: fast relocation to nucleolus and splicing speckles under hypoxia. Plant Cell 21:1592–1606
Lange TS, Gerbi SA (2000) Transient nucleolar localization Of U6 small nuclear RNA in Xenopus Laevis oocytes. Mol Biol Cell 11:2419–2428
Lee L, Davies SE, Liu JL (2009) The spinal muscular atrophy protein SMN affects Drosophila germline nuclear organization through the U body-P body pathway. Dev Biol 332:142–155
Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonné R, Lührmann R (2006) Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell 17:3221–3231
Liu JL, Gall JG (2007) U bodies are cytoplasmic structures that contain uridine-rich small nuclear ribonucleoproteins and associate with P bodies. Proc Natl Acad Sci USA 104:11655–11659
Liu JL, Buszczak M, Gall JG (2006a) Nuclear bodies in the Drosophila germinal vesicle. Chromosom Res 14:465–475
Liu JL, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG (2006b) The Drosophila melanogaster Cajal body. J Cell Biol 172:875–884
Majewska-Sawka A, Rodriguez-Garcia MI (1996) rRNA distribution during microspore development in anthers of Beta vulgaris L.: quantitative in situ hybridization analysis. J Cell Sci 109:859–866
Malatesta M, Zancanaro C, Martin TE, Chan EK, Amalric F, Lührmann R, Vogel P, Fakan S (1994) Cytochemical and immunocytochemical characterization of nuclear bodies during hibernation. Eur J Cell Biol 65:82–93
Massenet S, Pellizzoni L, Paushkin S, Mattaj IW, Dreyfuss G (2002) The SMN complex is associated with snRNPs throughout their cytoplasmic assembly pathway. Mol Cell Biol 22:6533–6541
Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK (2009) Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell 17:639–647
Mattaj IW (1986) Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell 46:905–911
Misteli T (2007) Beyond the sequence: cellular organization of genome function. Cell 128:787–800
Monneron A, Bernhard W (1969) Fine structural organization of the interphase cell nucleus of some mammalian cells. J Ultrastruct Res 27:266–288
Moreno Díaz de la Espina S, de la Espina S, Risueño MC, Medina FJ (1982) Ultrastructural cytochemistry and autoradiographic characterization of coiled bodies in the plant cell nucleus. Biol Cell 44:229–238
Navascues J, Berciano MT, Tucker KE, Lafarga M, Matera AG (2004) Targeting SMN to Cajal bodies and nuclear gems during neuritogenesis. Chromosoma 112:398–409
Niedojadło J, Górska-Brylass A (2003) New type of snRNP containing nuclear bodies in plant cells. Biol Cell 95:303–310
Pálfi Z, Bach M, Solymosy F, Lührmann R (1989) Purification of the major UsnRNPs from broad bean nuclear extracts and characterization of their protein constituents. Nucleic Acid Res 17:1445–1458
Parfenov VN, Pochukalina GN, Davis DS, Reinbold R, Schöler HR, Murti KG (2003) Nuclear distribution of Oct-4 transcription factor in transcriptionally active and inactive mouse oocytes and its relation to RNA polymerase II and splicing factors. J Cell Biochem 89:720–732
Pontes O, Pikaard CS (2008) siRNA and miNA processing: new functions for Cajal bodies. Curr Opin Genet 18:197–203
Ro-Choi TS (1999) Nuclear snRNA and nuclear function (discovery of 5′ cap structures in RNA). Crit Rev Eukaryot Gene Expr 9:107–158
Schisa JA, Pitt JN, Priess JR (2001) Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development 128:1287–1298
Seguí-Simarro JM, Bárány I, Suárez R, Fadón B, Testillano PS, Risueño MC (2006) Nuclear bodies domain changes with microspore reprogramming to embryogenesis. Eur J Histochem 50:35–44
Shaw PJ, Brown JW (2004) Plant nuclear bodies. Curr Opin Plant Biol 7:614–620
Sleeman JE, Ajuh P, Lamond AI (2001) snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci 114:4407–4419
Smoliński DJ, Kołowerzo A (2011) mRNA accumulation in the Cajal bodies of the diplotene larch microsporocyte. Chromosoma. doi: 10.1007/s00412-011-0339-4
Smoliński DJ, Niedojadło J, Noble A, Górska-Brylass A (2007) Additional nucleoli and NOR activity during meiotic prophase I in larch (Larix decidua Mill.). Protoplasma 232:109–120
Staněk D, Neugabauer KM (2006) The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma 115:343–354
Stanĕk D, Neugebauer KM (2004) Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J Cell Biol 166:1015–1025
Staněk D, Pridalová-Hnilicová J, Novotný I, Huranová M, Blazíková M, Wen X, Sapra AK, Neugebauer KM (2008) Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies. Mol Biol Cell 19:2534–2543
Straatman KR, Schel JH (2001) Distribution of splicing proteins and putative coiled bodies during pollen development and androgenesis in Brassica napus L. Protoplasma 216:181–200
Strzelecka M, Oates AC, Neugebauer KM (2010a) Dynamic control of Cajal body number during zebrafish embryogenesis. Nucleus 1:96–108
Strzelecka M, Trowitzsch S, Weber G, Lührmann R, Oates AC, Neugebauer KM (2010b) Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol 17:403–409
Testillano PS, Sánchez-Pina MA, Olmedilla A, Fuchs JP, Risueño MC (1993) Characterization of the interchromatin region as the nuclear domain containing snRNPs in plant cells. A cytochemical and immunoelectron microscopy study. Eur J Cell Biol 61:349–361
Tycowski KT, You Z-H, Graham PJ, Steitz JA (1998) Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol Cell 2:629–638
Visa N, Puvion-Dutilleul F, Harper F, Bachellerie JP, Puvion E (1993) Intranuclear distribution of poly(A) RNA determined by electron microscope in situ hybridization. Exp Cell Res 1:19–34
Will CL, Luhrmann R (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 13:290–301
Wróbel B, Smoliński DJ (2003) Coiled bodies in the meristematic cells of the root of Lupinus luteus L. Biologia Plantarum 46:223–232
Wu CH, Gall JG (1993) U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc Natl Acad Sci USA 90:6257–6259
Zienkiewicz K, Bednarska E (2009) snRNP: Rich nuclear bodies in Hyacinthus orientalis L. Microspores and developing pollen cells. Int J Cell Biol. doi:10.1155/2009/209303
Zienkiewicz K, Smoliński DJ, Bednarska E (2006) Distribution of poly(A) RNA and splicing machinery elements in mature Hyacinthus orientalis L. pollen grains and pollen tubes growing in vitro. Protoplasma 227:95–103
Zienkiewicz K, Zienkiewicz A, Smoliński DJ, Rafińska K, Świdziński M, Bednarska E (2008) Transcriptional state and distribution of poly(A) RNA and RNA polymerase II in differentiating Hyacinthus orientalis L. pollen grains. Sex Plant Reprod 21:233–245
Acknowledgments
We thank J. Niedojadło and E. Bednarska for critical reading of the manuscript. This work was supported by a grant from the Polish Ministry of Science and Higher Education no. N 303 799640.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
418_2011_861_MOESM1_ESM.tif
Supplementary Fig. S1 Immunoprecipitation with mouse anti-SmD (mAb 7.13). 1 Primary anti-SmD antibody probed with a secondary antibody, a control without sample. 2 Nuclear protein extracts immunoprecipitated with the anti-SmD antibody and probed with the same antibody. A single protein band with a mass of 55 kDa is visible. 3 Nuclear protein extracts immunoprecipitated with the anti-SmD antibody and probed with anti-m3G. A specific band with a molecular mass of 55 kDa can be observed. The bands at 60 and 28 kDa correspond to the heavy and light chains of the IgG. (TIFF 1789 kb)
418_2011_861_MOESM2_ESM.ppt
Supplementary Fig. S2 Immunoblot of Y12 antiserum probed with the total protein extract obtained from the anthers of Larix decidua. The protein bands with masses of about 30 and 14 kDa correspond to SmB/B and SmD1, respectively. (PPT 280 kb)
418_2011_861_MOESM3_ESM.ppt
Supplementary Fig. S3 Immunolocalization of m3G snRNA and Sm proteins (AF-ANA) during the premeiotic interphase using the double immunofluorescence labeling method. At the first stage, low levels of Sm proteins were observed in the cytoplasm, and Sm proteins were mainly present in a dispersed form (a). In nucleus, very low levels of Sm proteins and m3G snRNA were observed (a-c). In the second stage of the cycle, numerous fine clusters containing the Sm proteins appeared in the cytoplasm (e). Some larger cytoplasmic bodies containing both Sm proteins and m3G snRNA were observed (e–g). The major snRNP signal was observed near the nuclear border. In the next stage (i-l), when numerous cytoplasmic bodies were clearly visible in the cytoplasm, the proteins and snRNA appeared not only in the nuclear perimeter but also in tiny nuclear bodies (arrows). At the end of the cycle (m-p), when only fine single cytoplasmic bodies were visible in the cytoplasm (arrow), numerous smaller and larger nuclear bodies containing both Sm proteins and m3G snRNA can be observed in the nucleus. The corresponding DAPI images were collected using widefield fluorescence and deconvolution software (d, h, l, p). Bars, 10 μm. (PPT 1564 kb) (PPT 1564 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Smoliński, D.J., Wróbel, B., Noble, A. et al. Periodic expression of Sm proteins parallels formation of nuclear Cajal bodies and cytoplasmic snRNP-rich bodies. Histochem Cell Biol 136, 527–541 (2011). https://doi.org/10.1007/s00418-011-0861-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-011-0861-8