Abstract
Guanylin (GN) and uroguanylin (UGN), through activation of guanylyl cyclase C (GCC), serve to control intestinal fluid homeostasis. Both peptides are produced in the intestinal epithelium, but their cellular origin has not been fully charted. Using quantitative PCR and an improved in situ hybridization technique (RNAscope), we have assessed the expression of GN (Guca2a), UGN (Guca2b), and GCC (Gucy2c) in mouse intestine. In the crypts of Lieberkühn, expression of Guca2a and Guca2b was restricted to cells of secretory lineage, at the crypt’s base, and to a region above, previously identified as a common origin of cellular differentiation. In this compartment, comparatively uniform levels of Guca2a and Guca2b expression were observed throughout the length of the gut. In contrast, Guca2a and Guca2b expression in the villus–surface region was more variable, and reflected the distinct, but overlapping expression pattern observed previously. Accordingly, in jejunum and ileum, Guca2a and Guca2b were abundantly expressed by enterocytes, whereas in colon only Guca2a transcript was found in the surface region. In duodenum, only low levels of Guca2b transcript were observed in columnar cells, and Guca2a expression was restricted entirely to cells of the secretory lineage. Gucy2c was shown to be expressed relatively uniformly along the rostrocaudal and crypt–villus axes and was also found in the duodenal glands. Our study reveals novel aspects of the cellular localization of the GCC signaling axis that, apart from its role in the regulation of fluid balance, link it to pH regulation, cell cycle control, and host defense.
Similar content being viewed by others
Introduction
Signal transduction through the receptor–enzyme guanylyl cyclase C (GCC) serves to control intestinal fluid balance. Activation of the luminal, extracellular receptor domain by one of two locally produced peptides, guanylin (GN) and uroguanylin (UGN), leads to a surge in cellular cGMP levels, which prompts osmotic water transport to the intestinal lumen by protein kinase-mediated stimulation of anion secretion through the cystic fibrosis transmembrane conductance regulator (CFTR) anion channel and inhibition of sodium absorption through the sodium-proton exchanger type 3 (NHE3) (Vaandrager 2002). This pivotal role of GCC signaling in intestinal fluid balance is most poignantly illustrated by gain- and loss-of-function mutations in GUCY2C (encoding GCC), which have been shown to cause secretory diarrhea and intestinal obstruction, respectively (Romi et al. 2012; Fiskerstrand et al. 2012; Muller et al. 2015). Furthermore, GCC is activated by the (U)GN mimetic heat-stable toxin (STa) produced by enterotoxigenic Escherichia coli, a frequent cause of infectious diarrhea.
Apart from its role in the regulation of fluid homeostasis, GCC signaling may also regulate luminal pH. GCC stimulation strongly enhances duodenal bicarbonate secretion, and some data suggest that luminal acid may stimulate UGN release from duodenal enteroendocrine cells (Guba et al. 1996; Bengtsson et al. 2007; Kokrashvili et al. 2009; Joo et al. 1998; Rao et al. 2004; Singh et al. 2008). Evidently, in proximal intestine, such a feedback loop may aid the neutralization of the acid load entering from the stomach. In addition, it has become apparent that local bicarbonate production is crucial for the proper expansion of the mucins produced by goblet cells, and, consequently, the physiochemical properties of the mucus layer covering the epithelium along the entire tract (Garcia et al. 2009; Gustafsson et al. 2012). Possibly connected to its effect on mucin unfolding and fluid transport, loss of GCC signaling was shown to impair epithelial barrier function and innate host responses to bacterial pathogens (Mann et al. 2013; Han et al. 2011).
Lastly, GCC signaling may modulate the cell cycle in small intestinal and colonic epithelium. Mice deficient in either GN-, GCC-, or the cGMP-dependent protein kinase operating downstream of GCC (PRKG2), exhibit enhanced cell proliferation in the crypts and a reduction in the amount of cells of the secretory lineage (Li et al. 2007a; Steinbrecher et al. 2002; Wang et al. 2012b). It has also been suggested that loss of GCC signaling predisposes to tumor development (Li et al. 2007b).
The convergence of such apparently disparate physiological processes at the level of GCC suggests a strict compartmentalization of signaling routes. In this context, it is of interest that various distinct cell types within the intestinal epithelium have been proposed to produce GN and UGN. Mostly, these studies indicate production by cells from the secretory (granulocytic) lineage. In human small intestine, GN transcript was localized to the Paneth cells, whereas in the rat, GN transcript and protein were detected in goblet cells, and UGN transcript was found in enterochromaffin (EC) cells (De Sauvage et al. 1992; Cohen et al. 1995; Perkins et al. 1997; Date et al. 1996). In rat colon, GN was expressed by goblet cells, but also by the columnar cells of the surface epithelium, whereas in guinea pig, GN immunoreactivity was restricted to EC cells (Cohen et al. 1995; Cetin et al. 1994; Li et al. 1995). Finally, in mouse intestine, UGN transcript was localized exclusively to the villus, whereas GN transcript was found in both the villi and crypts of the small intestine and in the surface epithelium of colon (Whitaker et al. 1997). These disparate results may signify marked differences in the localization, and, perhaps, the physiological role of GN and UGN between species, and different anatomical regions. In addition, they may reflect the technical limitations of classical in situ hybridization and immunological methods.
Our study aimed to ascertain the expression pattern of GN, UGN, and GCC, using quantitative PCR and an improved in situ hybridization technique, which drastically improves sensitivity. While our study, in the main, corroborates previously reported patterns of GN, UGN, and GCC partitioning along the rostrocaudal axis of the intestine, these technical advances have allowed us to chart these patterns in more detail, revealing previously unappreciated differences in the expression pattern of duodenum versus jejunum, and the cell lineages involved in the production of GN, UGN, and GCC.
Materials and methods
Animals
Mice (FVB) were maintained in an environmentally controlled facility at the Erasmus MC, Rotterdam. All experiments were performed on animals, 8–16 weeks of age, and were approved of by the Ethical Committee for Animal Experiments of the Erasmus MC.
Tissue collection
Mice were anaesthetized (ketamine 100 mg/kg, xylazine 20 mg/kg; i.p.), and the intestinal tract was collected and flushed with ice-cold saline. All tissue was harvested between 12:00 and 14:00 h, to control for diurnal variations in gene expression.
For assessing the partitioning of transcript along the rostrocaudal axis of the intestinal tract, 6 equidistant segments (length: 0.5 cm) of small intestine and 2 segments of the colon were excised. The small intestine was sampled, starting 1 cm caudal to the pyloric sphincter and up to 1 cm proximal to the ileocecal valve. Colon was sampled at 1/3 and 2/3 of its entire length. Tissue was flash frozen in liquid nitrogen and stored at −80 °C.
For RNAscope analysis and lectin UEA1 (Ulex europaeus agglutinin 1) staining (see below), the excised intestine was flushed with ice-cold saline, cut open lengthwise, rolled into a Swiss roll, and immersed in PBS-buffered formalin (10 %) for 24 h at 4 °C. After fixation, tissue was embedded in paraffin, according to established protocols.
Quantitative polymerase chain reaction (qPCR)
Tissue was homogenized with a rotor–stator homogenizer in TRIzol reagent (Qiagen), and total RNA was extracted using the NucleoSpin RNA kit (Macherey–Nagel). After the integrity of the extracted RNA was verified by gel electrophoresis, cDNA was synthesized using the PrimeScript RT master mix (Takara Bio).
Quantitative PCR (primer sequences shown in Table S1) was performed on cDNA, using the SYBR Select Master Mix (Applied Bio System). Median values from assays performed in triplicate were used to determine the expression levels of Guca2a, Guca2b, and Gucy2c, relative to Gapdh.
In situ hybridization by RNAscope
Probes for detection of murine GN, UGN, and GCC transcripts were purchased from Advanced Cell Diagnostics (product code: 427996, 428006, 436596, respectively). Because, other than in conventional in situ hybridization, multiple independent probes have to hybridize to the target sequence in tandem in order for signal amplification to occur, RNAscope ensures selective amplification of target-specific signals (Wang et al. 2012a). RNAscope was performed according to the instructions of the manufacturer of the probes and the reagent kit (VS Reagent Kit 320600; Advanced Cell Diagnostics), on proteinase K (0.1 %, 5 min at 37 °C)-treated paraffin sections (5 µm). Detection of the ubiquitously expressed gene peptidylprolyl isomerase B (Ppib) served to ensure that tissue sections were correctly primed for probe hybridization (Advanced Cell Diagnostics, product code: 313919).
Histochemical detection of fucose glycoprotein
For detection of fucose glycoprotein-producing cells, sections used for RNAscope were re-hydrated in PBS, treated with a biotin blocking reagent (Dako), and incubated (1 h, at room temperature) with biotinylated lectin UEA1 (Vector Labs). Lectin UEA1-labeled carbohydrate moieties were visualized with a streptavidin-conjugated fluorescent probe (Alexa Fluor 594; Life Technologies). In negative controls, incubation with lectin UEA1 was omitted. Slides were mounted with ProLong Gold antifade reagent (Life Technologies) and stored at 4 °C.
Results
Partitioning of Guca2a, Guca2b, and Gucy2c transcripts along the rostrocaudal axis of the mouse intestinal tract
Quantitative PCR analysis showed that Guca2a transcript levels gradually increased along the rostrocaudal axis of the small intestine, and peaked in the proximal colon (Fig. 1a). In contrast, Guca2b transcript levels were low in colon. Guca2b levels were also low in the duodenum, but rose steeply along the rostrocaudal axis, and peaked in the middle to distal part of the small intestine (Fig. 1b). Gucy2c was expressed at much lower (>tenfold) levels than Guca2a or Guca2b and was partitioned more uniformly (Fig. 1c). Distribution of these transcripts was similar in male and female mice (not shown).
Partitioning of Guca2a (a), Guca2b (b), and Gucy2c (c) transcripts along the rostrocaudal axis of the mouse intestinal tract. Transcript levels in 6 equidistant sections of small intestine and 2 sections of colon (see diagram) were assessed by qPCR, using expression of Gapdh as a reference. Data depict mean ± standard error. N = 6
The RNAscope technique was used to visualize Guca2a, Guca2b, and Gucy2c transcripts in longitudinal sections of mouse intestine. This methodology employs up to 20 pairs of oligonucleotide probes per transcript, of which the paired probes need to hybridize in close proximity in order for signal amplification to occur. As a consequence, this technique provides a strongly improved signal-to-noise ratio compared with conventional in situ hybridization techniques (Wang et al. 2012a). The distribution pattern that emerged for these transcripts closely matched the expression profile assessed by qPCR analysis, i.e., the gradual increase of Guca2a and Guca2b expression from duodenum to the distal small intestine, the high expression of Guca2a, but low expression of Guca2b, in colon, and the comparatively low expression of Gucy2c (Fig. 2). This agreement strongly suggests specific hybridization of the probes. In addition, probe hybridization was restricted to intestinal epithelial cells, i.e., absent from underlying connective and muscle tissue, consistent with the discrete epithelial expression pattern of these genes. Our data also corroborate the previously observed high levels of focal Guca2b expression in regions of the colonic epithelium that border on lymphoid tissue (Fig. 2b colon section, Fig. 4j) (Whitaker et al. 1997). Specificity of the Guca2a probes was further corroborated by RNAscope performed on intestinal tissue of Guca2a null mice, in which only sparse punctuate staining was found in the nuclei (indicating weak hybridization with DNA) and the cytoplasmic region (indicating weak hybridization with truncated Guca2a transcripts; Fig. S1).
The observed rostrocaudal distribution patterns of GN, UGN, and GCC are congruent with those previously reported for rat intestine (Qian et al. 2000), but differ somewhat from those reported in a previous mouse study, in which the levels of Guca2b expression in duodenum and jejunum were shown to be similar (Whitaker et al. 1997). However, in this previous study, tissue from the proximal 3 cm of the mouse small intestine was pooled to assess duodenal expression, whereas we used a section between 1 and 1.5 cm distal to the pylorus. As is apparent from the RNAscope data (Fig. 2b), the expression level of Guca2b steeply increases within the first few centimeters of the small intestine, indicating that these apparent differences may be due largely to the different sampling strategies. Furthermore, it is conceivable that these differences reflect the effect of an unidentified environmental factor (i.e., diurnal rhythm, food intake, and composition) on duodenal Guca2b expression.
We did not detect Guca2a transcript in gastric or in pancreatic epithelia (Figs. 2a, 3), nor did we detect expression of Guca2b or Gucy2c in these tissues (Fig. 2b, c). This contrasts with previous observations on guinea pig, in which GN immunoreactivity was observed in pyloric EC cells (Cetin et al. 1994), and with observations on human and rat pancreatic tissue, in which GN, UGN, and GCC transcript and protein were detected (Kulaksiz et al. 2001; Kulaksiz and Cetin 2002). RT-PCR analysis confirmed that, compared to the gut, these glandular tissues contain only very low levels of these three transcripts (not shown). Such transcripts of low abundance may not be detectable by in situ hybridization, particularly in the pancreas, which contains high levels of ribonucleases (Azevedo-Pouly et al. 2014). However, arguing against significant mRNA loss prior to tissue fixation, we could readily detect Ppib transcript, in both the stomach and the pancreas (Fig. 3c, d). Therefore, we tentatively conclude that expression of the GCC signaling axis is low in mouse, compared to human pancreas. Interestingly, this apparent difference in the pancreatic expression of the GCC signaling axis between these species mirrors a previously observed disparity in the regulation of ductal anion and fluid secretion: Whereas in the human pancreas, the GCC signaling axis is thought to control the activity of the phosphorylation-regulated CFTR anion channel, anion and fluid secretion in the murine exocrine pancreas is principally governed by Ca2+-dependent anion channels, which operate independently of the GCC pathway (Winpenny et al. 1995; Kulaksiz et al. 2001).
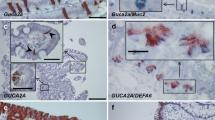
Partitioning of Guca2a and Ppib transcripts in the pyloric–duodenal region and in pancreas. a, b Guca2a expression was found in the duodenum (D), in cells at the base of the crypts of Lieberkühn, but was not detected in the gastric (G) or the duodenal glands (DG), or in the pancreas. c, d Detection of Ppib transcript in glandular epithelia in the pyloric–duodenal region and pancreas. IL islet of Langerhans
Partitioning of Guca2a and Guca2b transcripts along the crypt–villus and crypt–surface axis of the mouse small intestine and colon, respectively
Along the entire length of the small intestine, high Guca2a transcript levels were found in cell clusters at the base of the crypts and in solitary cells that line the lateral sides of the crypts (Fig. 4a–d). In the proximal small intestine, expression in the villus region was restricted to a distinct, sparsely distributed cell type (Fig. 4a). Very little expression was found in columnar, absorptive villus cells (enterocytes). However, in the proximal jejunum, expression of Guca2a in the villus became more prominent and was found in most columnar cells. Guca2a expression in the villus region further increased along the rostrocaudal axis, and distal jejunum and ileum showed abundant Guca2a expression in enterocytes (Fig. 4b, c). In colon, Guca2a transcript was found along the entire crypt–surface axis, but appeared to be most abundantly expressed by cells lining the lateral sides of the crypts and the surface. Strong expression was also found in cell clusters at the base of the crypts (Fig. 4d). Guca2a was also strongly expressed by the follicle-associated epithelium overlying lymphoid aggregates, in both small and large intestine (Fig. 4e).
Localization of Guca2a (a–d), Guca2b (f–i), and Gucy2c (k–n) transcripts in intestinal epithelium. e Guca2a expression in follicle-associated epithelium in proximal jejunum. j Guca2b expression in follicle-associated epithelium in colon. o Gucy2c expression in the region of transition from the gastric (G) to duodenal mucosa, with duodenal glands (DG) below. L lymphoid tissue
Like for Guca2a, Guca2b transcript was found in cells at the base and the lateral sides of the crypts, along the length of the small intestine (Fig. 4f–h). However, the level of cellular expression, and the amount of cells that expressed Guca2b in the crypts, appeared somewhat lower than for Guca2a. In the duodenum, Guca2b expression started to emerge at the base of the villi and involved the entire height of the villus in more distal sections of the small intestine (Fig. 4f–h). In colon, Guca2b expression was restricted to individual cells, or small clusters of cells, in the crypts (Fig. 4i). Although small in number, the level of expression per single cell was relatively high and comparable to the level in Guca2b-expressing cells in the crypts of the small intestine. As for Guca2a, we observed strong Guca2b expression in the follicle-associated epithelium overlying lymphoid tissue, in both small and large intestine (Fig. 2b colon section, Fig. 4j).
Partitioning of Gucy2c transcript along the crypt–villus and crypt–surface axis of the mouse small intestine and colon, respectively
Gucy2c was expressed relatively uniformly, in both the crypt and villus region of the epithelium (Fig. 4k–n). As for Guca2a and Guca2b, Gucy2c probe hybridization was restricted to intestinal epithelial cells. Expression in these cells was relatively uniform; only at the tip of the villi, expression seemed to wane slightly (similar was observed for Guca2a and Guca2b). No specific subset of cells was shown to be enriched in (or depleted of) Gucy2c transcript.
Further, we found Gucy2c transcripts in the duodenal glands (Fig. 4o). Previously, by autoradiographic demonstration of STa-specific binding sites in the intestinal mucosa, the presence of GCC in the duodenal glands of the North American opossum had been inferred (Krause et al. 1990). However, similar studies performed on human duodenal mucosa failed to demonstrate specific STa binding (Krause et al. 1994). Remarkably, the duodenal glands did not contain Guca2a or Guca2b transcript, indicating that local GCC activation relies on GN or UGN produced elsewhere.
Localization of Guca2a and Guca2b in cells of secretory lineage
In the small intestine, the Guca2a- and Guca2b-expressing cells at the base of the crypt were found to contain secretory granules and (in part) colocalized with Paneth cells (Fig. 5a, f). In contrast, Guca2a or Guca2b expression was not observed in the adjacent stem cells. A previous in situ hybridization study suggested that apparent Guca2b localization at the base of the crypts was a result of autofluorescence of the Paneth cells (Whitaker et al. 1997), but since RNAscope uses an enzymatic detection method, this cannot have confounded our observations. Moreover, our detection of Guca2a and Guca2b in Paneth cells is consistent with studies using a fluorescence-activated cell sorting technique to isolate specific epithelial cell lineages (Sato et al. 2011; Cash et al. 2006). This showed that Guca2a and Guca2b rank in the 90th percentile of most highly expressed genes in murine Paneth cells, and that Paneth cells are highly enriched in these transcripts, relative to stem cells (NCBI GEO datasets GSE5156 and GSE25109). In addition, these datasets reveal that Gucy2c is expressed at near-equal levels in Paneth and stem cells, congruent with the relatively uniform distribution of Gucy2c transcript in crypt epithelium (Fig. 4k–n).
Guca2a and Guca2b expression by cells of the secretory lineage in the duodenum and colon. a Guca2a expression in Paneth cells in duodenum. b Guca2a expression in cells at the base and the neck region of the crypts in colon. c Guca2a expression in a duodenal brush cell. d Guca2a expression in a duodenal goblet cell. e Costaining of Guca2a transcript (dark brown) and lectin UEA1-binding fucose glycoproteins (bright red) in duodenum. f Guca2b expression in Paneth cells in duodenum. g Guca2b expression in cells at the base of the crypts in colon. h Guca2b expression in a duodenal brush cell. No Guca2b transcript was observed in goblet cells. i, j Guca2b levels were generally low in columnar cells, but comparatively high in columnar cells adjoining goblet cells. B brush cell, G goblet cell, P Paneth cell
Guca2a and Guca2b transcripts were also found in cells at the base of the crypts in colon (Fig. 5b, g). These cells appear to occupy the same niche as the Paneth cells of the small intestine, suggesting they represent the cKit-positive Paneth-like cells identified previously (Rothenberg et al. 2012). In earlier studies, only Guca2a transcript had been detected in this compartment, possibly because the low sensitivity of conventional in situ hybridization techniques precluded detection of the less abundant Guca2b transcripts (Whitaker et al. 1997).
Lectin UEA1 staining confirmed that the cells at the base of the crypts and other Guca2a- and Guca2b-positive cells in the crypts and villus region of duodenum were of secretory lineage (Fig. 5e). Based on their numbers, localization, and typical shape, these Guca2a- and Guca2b-expressing cells in the villus appear to include brush (tuft) cells (Fig. 5c, h). Previously, a murine duodenal enteroendocrine cell-type secreting β-endorphin in response to acidic or hyperosmotic stimulation was identified (Kokrashvili et al. 2009). Interestingly, this cell type also contained UGN (but, based on immunological detection methods, not GN). In rat, apart from goblet cells, some enteroendocrine cell types also seemed to contain immunoreactive GN, although the specificity of the signal was questioned by the authors (Li et al. 1995). Collectively, these data strongly suggest that distinct brush cell populations coexist in the murine duodenal epithelium, expressing Guca2a or Guca2b.
In addition, Guca2a expression was found in a subset of goblet cells in the lower villus (Fig. 5d). This suggests that GN may be released concurrently with the mucin-containing granules produced by goblet cells. No Guca2b was found in goblet cells, but in the duodenum, where Guca2b expression is generally low, columnar cells that adjoined goblet cells seemed to have relatively (compared to more remote columnar cells) high Guca2b expression, suggesting that UGN release from these cells may play a role in goblet cell function (Fig. 5h–j). These findings are in line with studies on rat, which detected GN, but not UGN, in a subset of goblet cells of the rat small intestine and colon (Li et al. 1995; Cohen et al. 1995).
Discussion
An improved in situ hybridization technique (RNAscope) has allowed us to reveal novel aspects of the localization of the GCC signaling axis in the murine intestinal tract. We demonstrate strong expression of Guca2a and Guca2b transcripts in cells of secretory lineage, in particular Paneth cells located at the base of the crypts and chemosensory brush cells in the lower villus. Strikingly, the general pattern of Guca2a and Guca2b expression in this lineage is relatively uniform along the entire intestine: Even in the proximal part, which was thought to produce little GN, we observed prominent Guca2a expression in Paneth and brush cells, whereas in colon, which was thought to produce little UGN, robust Guca2b expression was found in Paneth-like cells at the base of the crypts.
In contrast to this relatively stable expression pattern in the crypts, expression in the villus–surface compartment showed more variation between anatomical regions, as expression of Guca2a and Guca2b extends to cells from the columnar (absorptive) lineage. It is chiefly the expression in this compartment that reiterates the previously reported pattern of GN and UGN distribution, i.e., a predominance of UGN in the proximal gut, and of GN in the colon (Qian et al. 2000; Whitaker et al. 1997). As has been noted before this pattern is consistent with the observation that GN is a relatively (in comparison with UGN) poor GCC ligand at the low pH prevailing in the proximal small intestine (Whitaker et al. 1997). Nevertheless, prominent expression of Guca2a is found in the crypts of the proximal small intestine, suggesting that the pH in this area may be maintained at more neutral values (aided by local bicarbonate secretion).
Along the length of the small intestine, Guca2a and Guca2b transcripts were found in Paneth cells, or, in case of the colon, in cells which appear to occupy the same position as these. Guca2a expression appeared more pronounced than Guca2b, but the localization pattern was similar. Previous in situ hybridization studies showed a similar distribution of Guca2a at the base of the crypts in jejunum and ileum of mice, although the exact cell types involved could not be ascertained (Whitaker et al. 1997). Guca2a and Guca2b expression by murine Paneth cells was also indicated in studies that employed cell sorting techniques to isolate epithelial cells representing different lineages (Sato et al. 2011; Cash et al. 2006). Furthermore, GN transcript was localized to human Paneth cells (De Sauvage et al. 1992).
It has been suggested that GN-stimulated fluid secretion emanating from the lower crypt may have a cleansing effect that supports the action of the bactericidal products of the Paneth cells (De Sauvage et al. 1992). In further support of innate immunity, GN and/or UGN release from Paneth cells may trigger bicarbonate secretion from neighboring CFTR-containing columnar cells, facilitating expansion of condensed mucins, and, consequently, the formation of the protective mucus layer covering the epithelium (Gustafsson et al. 2012; Garcia et al. 2009). In accordance with this notion, it has been shown that GCC signaling reduces susceptibility to infection by attaching/effacing bacterial pathogens (Mann et al. 2013). GN stored in goblet cells (and UGN in adjoining columnar cells) may serve a similar role. When released together with the mucin granules, a synchronized activation of GCC-dependent bicarbonate secretion from adjacent enterocytes (as goblet cells seem to contain little CFTR) may aid the proper expansion of the secreted condensed mucin granules. Consistent with this hypothesis, exocytosis from goblet cells is under cholinergic control, and it has been shown that cholinergic input also stimulates GN release in rat colon (Martin et al. 1999; Phillips 1992).
Apart from producing antibacterial products, Paneth cells also produce various factors that modulate stem cell function. It is conceivable that GN and UGN may also contribute to this modulation. In mice, it has been shown that GN, PRKG2, or GCC deficiency leads to enhanced crypt cell proliferation, and that GCC deficiency reduces goblet and Paneth cell numbers in mouse small intestine, whereas PRKG2 deficiency reduces goblet cell numbers in colon (Li et al. 2007a; Steinbrecher et al. 2002; Wang et al. 2012b). These results suggest that GCC signaling directs differentiation to a secretory lineage. Indeed, PRKG2 has been shown to control the activity of the transcription factor Sox9, which is a key determinant of Paneth cell formation (Mori-Akiyama et al. 2007; Swartling et al. 2009; Chikuda et al. 2004). Because of their close contact to stem cells and their progenitors, the Paneth cells are a likely source of the ligands that control GCC activity in this niche. Consistent with this model, both our present data and previous studies (NCBI GEO datasets GSE5156 and GSE25109) indicate that Gucy2c expression in the crypts extends to the stem cells, whereas Guca2a and Guca2b expression is restricted to cells of the secretory lineage (Sato et al. 2011; Cash et al. 2006).
In duodenum, prominent expression of Guca2a and Guca2b in the villi is mostly confined to just the few cells of secretory lineage. Apart from Paneth and goblet cells, this includes brush cells. UGN, but not GN, has previously been localized to murine brush cells (Kokrashvili et al. 2009). Brush cells are known to monitor the composition of the lumen, and it was shown that this particular subset of UGN-containing duodenal brush cells secretes β-endorphin into the lumen upon acidic or hypertonic stimulation (Kokrashvili et al. 2009; Ronnestad et al. 2014). We propose that the UGN stored in this cell type may also be released upon exposure to these luminal stimuli. In support, an increase in bicarbonate secretion was observed in mouse duodenum upon lowering of the luminal pH, which may be attributed to endogenous (U)GN release, as GN and UGN are known to strongly stimulate duodenal bicarbonate secretion (Singh et al. 2008; Guba et al. 1996; Joo et al. 1998). This assumption is also consistent with the observation that (I) luminal hypertonicity stimulates the release of GN and UGN and inhibits NHE3-mediated sodium absorption (Gawenis et al. 2003; Steinbrecher et al. 2001), and (II) enteral salt loading may induce intra-arterial UGN release from the intestinal mucosa (to stimulate renal natriuresis) (Lorenz et al. 2003). Collectively, these data suggest that GN and UGN are released from distinct subsets of brush cells upon acidic or hypertonic stimulation, and that their release serves to regulate luminal pH and osmolarity.
We observed expression of Gucy2c, but not of Guca2a or Guca2b, in the duodenal glands. This GCC pool may be targeted by GN and/or UGN released into the duodenal lumen (although diffusion of peptides into the glands is likely to be opposed by secretory fluid movement) and/or by systemic GN and UGN. In support of the latter option, intra-arterial GN or UGN has been shown to trigger duodenal bicarbonate secretion (Bengtsson et al. 2007). Indeed, systemic UGN has been shown to elicit additional GCC-dependent effects, notably in neuroendocrine tissues (Valentino et al. 2011; Gong et al. 2011). In further support of such actions in the gut, it was shown that contraluminal administration of STa to duodenal enteroids provoked GCC-mediated inhibition of Na+, H+ antiport activity, strongly suggesting that such oligopeptides are translocated across the epithelium, although the presence of GCC at the basolateral aspect of polarized intestinal epithelial cells cannot be excluded completely (Foulke-Abel et al. 2016).
Distal from the duodenum, prominent Guca2a and Guca2b expression is found in the columnar (absorptive) cells of the villus. Strong expression of Guca2a and Guca2b in the villus has been observed before in mice, but not in rat or guinea pig (Cetin et al. 1994; Steinbrecher et al. 2002; Perkins et al. 1997; De Sauvage et al. 1992; Wang et al. 2012b). Thus, the increase in total expression of Guca2a and Guca2b distal from the duodenum can be mainly attributed to expression initiated in enterocytes. This expression pattern is similar to that of the transcription factor Cdx2, suggesting that this, or other intestine-specific transcription factors, controls Guca2a and Guca2b expression in enterocytes (Fang et al. 2006). GN and/or UGN released in this region is unlikely to enter the crypts, which contain most of the CFTR-mediated anion-secretory capacity, and a function in the villus compartment seems indicated. Apart from activating a subpool of CFTR, GN and UGN may limit sodium absorption through NHE3, which is highly expressed in mature enterocytes and is inhibited through cGMP-dependent protein phosphorylation (Foulke-Abel et al. 2016). Assuming that peptides released at the surface of the villi would diffuse more rapidly into the bulk luminal content than those secreted in the crypts, local GCC activity and ion transport may simply be controlled by the level to which its ligands are diluted upon their release; GCC stimulation would peak at a low level of hydration, when a high concentration of its ligands is maintained at the epithelial surface. This may constitute a relatively straightforward mechanism to maintain fluid homeostasis, which does not so much rely on a stimulus-controlled release of GN and UGN, but on a continual secretion from columnar cells in the villus. It is of interest that the combined Guca2a and Guca2b expression in this compartment is particularly high in the distal small intestine, a region which appears especially prone to dehydration and obstruction caused by GCC deficiency (Romi et al. 2012).
In the colon, strong Guca2a expression was found in cells positioned at the lateral sides of the crypts. At the base of the crypts, expression was markedly lower and confined to a Paneth-like cell type (Rothenberg et al. 2012). The distal colon in particular contains high numbers of goblet cells, and these also appeared to express Guca2a. As in the small intestine, GN release from goblet cells may play a role in mucin expansion. Guca2b expression was low in colon, but was retained in solitary cells in the crypts. Strikingly, high Guca2a and Guca2b expression was found in those parts of the epithelium that were in contact with lymphoid tissue. This particular expression pattern has been observed before for Guca2b in the ileocecal region of mice (Whitaker et al. 1997). Because the follicle-associated epithelium also shows strong expression of genes encoding specific ion channels, i.e., Cftr, Clca2, Kcnj15, and Kcna1 (Kobayashi et al. 2012), and we also found weak expression of Gucy2c in this region, it is conceivable that GN and UGN regulate ion transport across the follicle-associated epithelium through autocrine signaling.
In summary, the present study charts Guca2a, Guca2b, and Gucy2c expression along the entire murine intestinal tract. Whereas Gucy2c expression is relatively uniform along the rostrocaudal and crypt–villus axes, strong regional differences exist in the expression of its ligands. These differences mostly reflect varying levels of expression in the upper regions of the epithelium. Gucy2c expression was found in the duodenal glands, suggesting it serves a role in UGN/GN-dependent duodenal bicarbonate secretion. Further, we show distinct expression of Guca2a and Guca2b in Paneth cells and brush cells, suggesting that GN and UGN play a role in chemoreception, stem cell proliferation, and host defense.
References
Azevedo-Pouly AC, Elgamal OA, Schmittgen TD (2014) RNA isolation from mouse pancreas: a ribonuclease-rich tissue. J Vis Exp 90:e51779
Bengtsson MW, Jedstedt G, Flemstrom G (2007) Duodenal bicarbonate secretion in rats: stimulation by intra-arterial and luminal guanylin and uroguanylin. Acta Physiol (Oxf) 191:309–317
Cash HL, Whitham CV, Behrendt CL, Hooper LV (2006) Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313:1126–1130
Cetin Y, Kuhn M, Kulaksiz H, Adermann K, Bargsten G, Grube D, Forssmann WG (1994) Enterochromaffin cells of the digestive system: cellular source of guanylin, a guanylate cyclase-activating peptide. Proc Natl Acad Sci USA 91:2935–2939
Chikuda H, Kugimiya F, Hoshi K, Ikeda T, Ogasawara T, Shimoaka T, Kawano H, Kamekura S, Tsuchida A, Yokoi N, Nakamura K, Komeda K, Chung UI, Kawaguchi H (2004) Cyclic GMP-dependent protein kinase II is a molecular switch from proliferation to hypertrophic differentiation of chondrocytes. Genes Dev 18:2418–2429
Cohen MB, Witte DP, Hawkins JA, Currie MG (1995) Immunohistochemical localization of guanylin in the rat small intestine and colon. Biochem Biophys Res Commun 209:803–808
Date Y, Nakazato M, Yamaguchi H, Miyazato M, Matsukura S (1996) Tissue distribution and plasma concentration of human guanylin. Intern Med 35:171–175
De Sauvage FJ, Keshav S, Kuang WJ, Gillett N, Henzel W, Goeddel DV (1992) Precursor structure, expression, and tissue distribution of human guanylin. Proc Natl Acad Sci USA 89:9089–9093
Fang R, Olds LC, Sibley E (2006) Spatio-temporal patterns of intestine-specific transcription factor expression during postnatal mouse gut development. Gene Expr Patterns 6:426–432
Fiskerstrand T, Arshad N, Haukanes BI, Tronstad RR, Pham KD, Johansson S, Havik B, Tonder SL, Levy SE, Brackman D, Boman H, Biswas KH, Apold J, Hovdenak N, Visweswariah SS, Knappskog PM (2012) Familial diarrhea syndrome caused by an activating GUCY2C mutation. N Engl J Med 366:1586–1595
Foulke-Abel J, In J, Yin J, Zachos NC, Kovbasnjuk O, Estes MK, De Jonge H, Donowitz M (2016) Human enteroids as a model of upper small intestinal ion transport physiology and pathophysiology. Gastroenterology 150:638–649
Garcia MA, Yang N, Quinton PM (2009) Normal mouse intestinal mucus release requires cystic fibrosis transmembrane regulator–dependent bicarbonate secretion. J Clin Invest 119:2613–2622
Gawenis LR, Franklin CL, Simpson JE, Palmer BA, Walker NM, Wiggins TM, Clarke LL (2003) cAMP inhibition of murine intestinal Na+/H+ exchange requires CFTR-mediated cell shrinkage of villus epithelium. Gastroenterology 125:1148–1163
Gong R, Ding C, Hu J, Lu Y, Liu F, Mann E, Xu F, Cohen MB, Luo M (2011) Role for the membrane receptor guanylyl cyclase-C in attention deficiency and hyperactive behavior. Science 333:1642–1646
Guba M, Kuhn M, Forssmann WG, Classen M, Gregor M, Seidler U (1996) Guanylin strongly stimulates rat duodenal HCO3 − secretion: proposed mechanism and comparison with other secretagogues. Gastroenterology 111:1558–1568
Gustafsson JK, Ermund A, Ambort D, Johansson ME, Nilsson HE, Thorell K, Hebert H, Sjovall H, Hansson GC (2012) Bicarbonate and functional CFTR channel are required for proper mucin secretion and link cystic fibrosis with its mucus phenotype. J Exp Med 209:1263–1272
Han X, Mann E, Gilbert S, Guan Y, Steinbrecher KA, Montrose MH, Cohen MB (2011) Loss of guanylyl cyclase C (GCC) signaling leads to dysfunctional intestinal barrier. PLoS One 6:e16139
Joo NS, London RM, Kim HD, Forte LR, Clarke LL (1998) Regulation of intestinal Cl− and HCO3 − secretion by uroguanylin. Am J Physiol 274:G633–G644
Kobayashi A, Donaldson DS, Kanaya T, Fukuda S, Baillie JK, Freeman TC, Ohno H, Williams IR, Mabbott NA (2012) Identification of novel genes selectively expressed in the follicle-associated epithelium from the meta-analysis of transcriptomics data from multiple mouse cell and tissue populations. DNA Res 19:407–422
Kokrashvili Z, Rodriguez D, Yevshayeva V, Zhou H, Margolskee RF, Mosinger B (2009) Release of endogenous opioids from duodenal enteroendocrine cells requires Trpm5. Gastroenterology 137:598–606
Krause WJ, Freeman RH, Fort LR (1990) Autoradiographic demonstration of specific binding sites for E. coli enterotoxin in various epithelia of the North American opossum. Cell Tissue Res 260:387–394
Krause WJ, Cullingford GL, Freeman RH, Eber SL, Richardson KC, Fok KF, Currie MG, Forte LR (1994) Distribution of heat-stable enterotoxin/guanylin receptors in the intestinal tract of man and other mammals. J Anat 184:407–417
Kulaksiz H, Cetin Y (2002) The electrolyte/fluid secretion stimulatory peptides guanylin and uroguanylin and their common functional coupling proteins in the rat pancreas: a correlative study of expression and cell-specific localization. Pancreas 25:170–175
Kulaksiz H, Schmid A, Honscheid M, Eissele R, Klempnauer J, Cetin Y (2001) Guanylin in the human pancreas: a novel luminocrine regulatory pathway of electrolyte secretion via cGMP and CFTR in the ductal system. Histochem Cell Biol 115:131–145
Li Z, Taylor-Blake B, Light AR, Goy MF (1995) Guanylin, an endogenous ligand for C-type guanylate cyclase, is produced by goblet cells in the rat intestine. Gastroenterology 109:1863–1875
Li P, Lin JE, Chervoneva I, Schulz S, Waldman SA, Pitari GM (2007a) Homeostatic control of the crypt-villus axis by the bacterial enterotoxin receptor guanylyl cyclase C restricts the proliferating compartment in intestine. Am J Pathol 171:1847–1858
Li P, Schulz S, Bombonati A, Palazzo JP, Hyslop TM, Xu Y, Baran AA, Siracusa LD, Pitari GM, Waldman SA (2007b) Guanylyl cyclase C suppresses intestinal tumorigenesis by restricting proliferation and maintaining genomic integrity. Gastroenterology 133:599–607
Lorenz JN, Nieman M, Sabo J, Sanford LP, Hawkins JA, Elitsur N, Gawenis LR, Clarke LL, Cohen MB (2003) Uroguanylin knockout mice have increased blood pressure and impaired natriuretic response to enteral NaCl load. J Clin Invest 112:1244–1254
Mann EA, Harmel-Laws E, Cohen MB, Steinbrecher KA (2013) Guanylate cyclase C limits systemic dissemination of a murine enteric pathogen. BMC Gastroenterol 13:135
Martin S, Adermann K, Forssmann WG, Kuhn M (1999) Regulated, side-directed secretion of proguanylin from isolated rat colonic mucosa. Endocrinology 140:5022–5029
Mori-Akiyama Y, van den Born M, van Es JH, Hamilton SR, Adams HP, Zhang J, Clevers H, de Crombrugghe B (2007) SOX9 is required for the differentiation of Paneth cells in the intestinal epithelium. Gastroenterology 133:539–546
Muller T, Rasool I, Heinz-Erian P, Mildenberger E, Hulstrunk C, Muller A, Michaud L, Koot BG, Ballauff A, Vodopiutz J, Rosipal S, Petersen BS, Franke A, Fuchs I, Witt H, Zoller H, Janecke AR, Visweswariah SS (2015) Congenital secretory diarrhoea caused by activating germline mutations in GUCY2C. Gut. doi:10.1136/gutjnl-2015-309441
Perkins A, Goy MF, Li Z (1997) Uroguanylin is expressed by enterochromaffin cells in the rat gastrointestinal tract. Gastroenterology 113:1007–1014
Phillips TE (1992) Both crypt and villus intestinal goblet cells secrete mucin in response to cholinergic stimulation. Am J Physiol 262:G327–G331
Qian X, Prabhakar S, Nandi A, Visweswariah SS, Goy MF (2000) Expression of GC-C, a receptor-guanylate cyclase, and its endogenous ligands uroguanylin and guanylin along the rostrocaudal axis of the intestine. Endocrinology 141:3210–3224
Rao SP, Sellers Z, Crombie DL, Hogan DL, Mann EA, Childs D, Keely S, Sheil-Popoulo M, Giannella RA, Barrett KE, Isenberg JI, Pratha VS (2004) A role for guanylate cyclase C (GC-C) in acid-stimulated duodenal mucosal bicarbonate secretion. Am J Physiol 286:G95–G101
Romi H, Cohen I, Landau D, Alkrinawi S, Yerushalmi B, Hershkovitz R, Newman-Heiman N, Cutting GR, Ofir R, Sivan S, Birk OS (2012) Meconium ileus caused by mutations in GUCY2C, encoding the CFTR-activating guanylate cyclase 2C. Am J Hum Genet 90:893–899
Ronnestad I, Akiba Y, Kaji I, Kaunitz JD (2014) Duodenal luminal nutrient sensing. Curr Opin Pharmacol 19:67–75
Rothenberg ME, Nusse Y, Kalisky T, Lee JJ, Dalerba P, Scheeren F, Lobo N, Kulkarni S, Sim S, Qian D, Beachy PA, Pasricha PJ, Quake SR, Clarke MF (2012) Identification of a cKit(+) colonic crypt base secretory cell that supports Lgr5(+) stem cells in mice. Gastroenterology 142(1195–1205):e1196
Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H (2011) Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469:415–418
Singh AK, Sjoblom M, Zheng W, Krabbenhoft A, Riederer B, Rausch B, Manns MP, Soleimani M, Seidler U (2008) CFTR and its key role in in vivo resting and luminal acid-induced duodenal HCO3 − secretion. Acta Physiol (Oxf) 193:357–365
Steinbrecher KA, Mann EA, Giannella RA, Cohen MB (2001) Increases in guanylin and uroguanylin in a mouse model of osmotic diarrhea are guanylate cyclase C-independent. Gastroenterology 121:1191–1202
Steinbrecher KA, Wowk SA, Rudolph JA, Witte DP, Cohen MB (2002) Targeted inactivation of the mouse guanylin gene results in altered dynamics of colonic epithelial proliferation. Am J Pathol 161:2169–2178
Swartling FJ, Ferletta M, Kastemar M, Weiss WA, Westermark B (2009) Cyclic GMP-dependent protein kinase II inhibits cell proliferation, Sox9 expression and Akt phosphorylation in human glioma cell lines. Oncogene 28:3121–3131
Vaandrager AB (2002) Structure and function of the heat-stable enterotoxin receptor/guanylyl cyclase C. Mol Cell Biochem 230:73–83
Valentino MA, Lin JE, Snook AE, Li P, Kim GW, Marszalowicz G, Magee MS, Hyslop T, Schulz S, Waldman SA (2011) A uroguanylin-GUCY2C endocrine axis regulates feeding in mice. J Clin Invest 121:3578–3588
Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y (2012a) RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 14:22–29
Wang R, Kwon IK, Thangaraju M, Singh N, Liu K, Jay P, Hofmann F, Ganapathy V, Browning DD (2012b) Type 2 cGMP-dependent protein kinase regulates proliferation and differentiation in the colonic mucosa. Am J Physiol 303:G209–G219
Whitaker TL, Witte DP, Scott MC, Cohen MB (1997) Uroguanylin and guanylin: distinct but overlapping patterns of messenger RNA expression in mouse intestine. Gastroenterology 113:1000–1006
Winpenny JP, Verdon B, McAlroy HL, Colledge WH, Ratcliff R, Evans MJ, Gray MA, Argent BE (1995) Calcium-activated chloride conductance is not increased in pancreatic duct cells of CF mice. Pflugers Arch 430:26–33
Funding
This work was supported by the Dutch Cystic Fibrosis Foundation (NCFS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Ron Smits and Marcel J. C. Bijvelds have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ikpa, P.T., Sleddens, H.F.B.M., Steinbrecher, K.A. et al. Guanylin and uroguanylin are produced by mouse intestinal epithelial cells of columnar and secretory lineage. Histochem Cell Biol 146, 445–455 (2016). https://doi.org/10.1007/s00418-016-1453-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-016-1453-4








