Abstract
Opium poppy (Papaver somniferum) is one of the world’s oldest medicinal plants and remains the only commercial source for the narcotic analgesics morphine, codeine and semi-synthetic derivatives such as oxycodone and naltrexone. The plant also produces several other benzylisoquinoline alkaloids with potent pharmacological properties including the vasodilator papaverine, the cough suppressant and potential anticancer drug noscapine and the antimicrobial agent sanguinarine. Opium poppy has served as a model system to investigate the biosynthesis of benzylisoquinoline alkaloids in plants. The application of biochemical and functional genomics has resulted in a recent surge in the discovery of biosynthetic genes involved in the formation of major benzylisoquinoline alkaloids in opium poppy. The availability of extensive biochemical genetic tools and information pertaining to benzylisoquinoline alkaloid metabolism is facilitating the study of a wide range of phenomena including the structural biology of novel catalysts, the genomic organization of biosynthetic genes, the cellular and sub-cellular localization of biosynthetic enzymes and a variety of biotechnological applications. In this review, we highlight recent developments and summarize the frontiers of knowledge regarding the biochemistry, cellular biology and biotechnology of benzylisoquinoline alkaloid biosynthesis in opium poppy.
Similar content being viewed by others
Introduction
Opium poppy (Papaver somniferum) is an ancient medicinal plant that remains the only commercial source of the narcotic analgesics morphine, codeine and semi-synthetic analogs including oxycodone, hydrocodone, buprenorphine and naltrexone (Berényi et al. 2009). Other pharmaceutically important benzylisoquinoline alkaloids (BIAs) found in opium poppy include the antimicrobial agent sanguinarine, the muscle relaxant papaverine and the cough-suppressant and potential anticancer drug noscapine. The ~2,500 known BIAs represent a diverse group of nitrogen-containing specialized metabolites found mainly in the basal eudicot order Ranunculales, in particular the families Papaveraceae, Ranunculaceae, Berberidaceae and Menispermaceae (Liscombe et al. 2005). Many BIAs of pharmaceutical importance possess one or more chiral centers, which preclude chemical synthesis as an economically viable option for commercial production. In addition to its pharmaceutical importance, opium poppy is also cultivated as an illicit source of morphine, which is easily converted to O,O-diacetylmorphine, or heroin. Opium poppy is a classic example of dual-use technology whereby compounds of both great positive and negative value to humanity are produced.
The biochemistry and physiology of BIA metabolism in opium poppy have long been investigated. Major advances were achieved through (1) the application of radiotracer techniques in the 1960s, (2) enzyme isolation and characterization methodologies in the 1980s and (3) recombinant DNA technologies in the 1990s (Hagel and Facchini 2013). The recent applications of next-generation DNA sequencing systems and powerful functional genomics methods have expedited the rate of discovery. The cumulative knowledge acquired over the past half-century provides a remarkable platform for understanding BIA metabolism in opium poppy. This review summarizes recent developments and current frontiers in our understanding that define the status of opium poppy as a model system for the study of alkaloid biosynthesis in plants.
Biosynthesis of the major alkaloids in opium poppy
Many reactions from a limited number of enzyme families
Enzymes involved in BIA metabolism belong to a relatively limited number of protein families, including cytochromes P450 (CYPs), S-adenosylmethionine-dependent O- and N-methyltransferases, four distinct groups of NADPH-dependent dehydrogenases/reductases, FAD-linked oxidoreductases (FADOXs), and more restricted numbers of acetyl-CoA-dependent O-acetyltransferases, 2-oxoglutarate/Fe(II)-dependent dioxygenases (ODDs) and carboxylesterases. The monophylogeny of BIA biosynthesis in the Ranunculales (Liscombe et al. 2005) facilitates the identification of novel biosynthetic genes based on amino acid sequence similarity with previously characterized enzymes. Additional enzyme families, including UDP-dependent glucosyltransferases, are undoubtedly involved in uncharacterized aspects of alkaloid metabolism based on the occurrence of signature metabolites, such as glucosylated BIAs produced by these enzymes (Liscombe et al. 2009).
The remarkable structural diversity of BIAs is achieved initially through modification of the 1-benzylisoquinoline backbone, but multiplies extensively by the addition of various functional groups. Remodeling of the 1-benzylisoquinoline scaffold is mediated by oxidative enzymes (i.e. CYPs, FADOXs and ODDs). CYPs catalyze a wide variety of reactions ranging from hydroxylations to C–C and C–O couplings and are responsible for the formation of most BIA structural subgroups with the exception of protoberberines. In BIA metabolism, FADOXs have only been implicated in the formation of C–C and C–N bonds, the most noteworthy being the berberine bridge enzyme (BBE), which generates the protoberberine scaffold (Dittrich and Kutchan 1991; Facchini et al. 1996; Winkler et al. 2006). In contrast, FADOX homologs appear to only catalyze the formation of C–O bonds in the formation of non-alkaloid specialized metabolites in other plants (Custers et al. 2004; Sirikantaramas et al. 2004). Although ODDs are known to perform a variety of hydroxylations, epoxidations and desaturations in a variety of plant metabolic pathways (Prescott and John 1996), enzymes from this family have only been shown to catalyze the dealkylation of certain BIA subgroups (Hagel and Facchini 2010; Farrow and Facchini 2013).
A collection of diverse O-methytransferases catalyze the regiospecific transfer of a methyl group from S-adenosylmethionine to a free hydroxyl moiety on a variety of BIA subgroups, but appear to primarily target 1-benzylisoquinolines and protoberberines (Dang and Facchini 2012). N-methyltransferases involved in BIA biosynthesis display substantial sequence similarity and catalyze the transfer of a methyl group to the nitrogen on the isoquinoline ring. Despite the occurrence of a similar S-adenosylmethionine binding domain, it is still not known whether O- and N-methyltransferases have a common ancestor (Ibrahim and Muzac 2000).
(S)-Reticuline
BIA biosynthesis begins with the condensation of two l-tyrosine derivatives, 4-hydroxyphenylacetaldehyde (4HPAA) and dopamine, via decarboxylation, meta-hydroxylation and transamination yielding the precursor to all other BIAs, (S)-norcoclaurine (Fig. 1). In the formation of 4HPAA, l-tyrosine can undergo transamination by l-tyrosine aminotransferase (TyrAT) (Lee and Facchini 2011) and subsequent decarboxylation by an unidentified enzyme isolated as 4-hydroxyphenylpyruvate decarboxylase (4HPPDC) (Rueffer and Zenk 1987). meta-Hydroxylation of l-tyrosine and/or tyramine by another uncharacterized enzyme (referred to as 3′OHase in Fig. 1) yields l-dihydroxyphenylalanine (DOPA) and dopamine, respectively. Alternatively, l-tyrosine and DOPA are converted to tyramine and dopamine by tyrosine decarboxylase (TYDC) (Facchini and De Luca 1994). The stereoselective Pictet–Spengler condensation of (S)-norcoclaurine from 4HPAA and dopamine is catalyzed by norcoclaurine synthase (NCS), which is a member of the pathogenesis-related (PR)10/Bet v 1 protein family (Liscombe et al. 2005; Samanani et al. 2004; Lee and Facchini 2010). In addition to a PR10-type NCS, a second enzyme (CjNCS1) sharing similarity with ODDs was reported to catalyze the formation of (S)-norcoclaurine in Coptis japonica (Minami et al. 2007). However, the actual physiological role of this enzyme is not known.
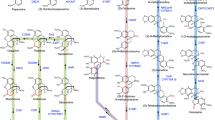
Biosynthetic pathways leading to major benzylisoquinoline alkaloids accumulating in opium poppy and other plant species. Pathways: pink 1-benzylisoquinoline; green, morphinan; dark blue, papaverine; purple, phthalideisoquinoline; orange, benzo[c]phenanthridine; light blue, protoberberine; brown, bisbenzylisoquinoline; dark pink, aporphine. Enzymes for which corresponding genes have been isolated from opium poppy or from other plants are shown in green and blue, respectively. Enzymes for which corresponding genes have not been isolated are shown in black. Chemical conversions catalyzed by each enzyme are shown in red. TYDC tyrosine/DOPA decarboxylase, 3OHase tyrosine/tyramine 3-hydroxylase, 4HPPDC 4-hydroxyphenylpuruvate decarboxylase, NCS norcoclaurine synthase, 6OMT norcoclaurine 6-O-methyltransferase, CNMT coclaurine N-methyltransferase, NMCH N-methylcoclaurine 3′-hydroxylase, 4′OMT2 3′-hydroxyl-N-methylcoclaurine 4′-O-methyltransferase (isoform 2), BBE berberine bridge enzyme, SOMT1 scoulerine 9-O-methyltransferase, CAS canadine synthase, TNMT tetrahydroprotoberberine N-methyltransferase, CYP82Y1 N-methylcanadine 1-hydroxylase, NOS noscapine synthase, STOX (S)-tetrahydroxyprotoberberineoxidase, CoOMT columbamine O-methyltransferase, CFS cheilanthifoline synthase, SPS stylopine synthase, MSH N-methylstylopine 14-hydroxylase, P6H protopine 6-hydroxylase, DBOX dihydrosanguinarine oxidase, SanR sanguinarine reductase, SalSyn salutaridine synthase, SalR salutaridine reductase, SalAT salutaridinol 7-O-acetyltransferase, T6ODM thebaine 6-O-demethylase, COR codeinone reductase, CODM codeine O-demethylase, N7OMT norreticuline 7-O-methyltransferase, 3′OHase uncharacterized 3′-hydroxylase, 3′OMT uncharacterized 3′-O-methyltransferase
Many BIAs are derived from the central pathway intermediate (S)-reticuline, which is formed via 3′-hydroxylation and a series of O- and N-methylations (Fig. 1). The conversion order was inferred by the substrate preference of the relevant enzymes although recent evidence suggests that the pathway is not linear, but rather operates as a grid (Desgagné-Penix and Facchini 2012). (S)-Norcoclaurine is first converted by norcoclaurine-6-O-methyltransferase (6OMT) to (S)-coclaurine (Morishige et al. 2000; Ounaroon et al. 2003), which is accepted by coclaurine N-methyltransferase (CNMT) (Choi et al. 2001). N-Methylcoclaurine is hydroxylated by (S)-N-methylcoclaurine 3′-hydroxylase (NMCH) (Pauli and Kutchan 1998; Frick et al. 2007) to 3′-hydroxy-N-methylcoclaurine, which is converted by 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase (4′OMT) (Morishige et al. 2000) to (S)-reticuline. Interestingly, only NMCH has been reported to show strict substrate and stereoisomer specificity, accepting (S)-N-methylcoclaurine and not the corresponding (R)-N-methylcoclaurine or N-desmethyl compounds. In contrast, the O- and N-methyltransferases typically accept a variety of (R)- and (S)-tetrahydroisoquinolines. Reticuline can be further methylated by reticuline 7-O-methyltransferase (7OMT) (Ounaroon et al. 2003) yielding laudanine, which can be fully O-methylated to laudanosine by an unidentified 3′-O-methyltransferase (indicated as 3′OMT in Fig. 1).
Papaverine
Two routes for papaverine biosynthesis have been proposed: (1) an N-methyl pathway involving (S)-reticuline and the N-demethylation of an unspecified intermediate by a hypothetical enzyme and (2) an N-desmethyl pathway involving (S)-norreticuline and precluding the requirement for N-demethylation (Fig. 1). The efficient incorporation of radiolabeled N-desmethyl compounds (Brochmann-Hanssen et al. 1975; Uprety et al. 1975) and the identification of a norreticuline 7-O-methyltransferase (N7OMT), which accepts only norreticuline yielding norlaudanine (Pienkny et al. 2009), provides biochemical support for the N-desmethyl pathway. A recent study using mass spectrometry and heavy stable-isotope labeling of (S)-reticuline, (S)-laudanine and (S)-laudanosine suggested that N-methylated 1-benzylisoquinolines are incorporated into papaverine at low levels (Han et al. 2010), providing some support for the N-methyl pathway. However, most of the labeled (S)-reticuline was incorporated into morphinan alkaloids.
The use of virus-induced gene silencing (VIGS) further demonstrated that the major route to papaverine is likely through the N-desmethyl pathway and showed that norreticuline and other N-desmethyled 1-benzylisoquinolines are natural metabolites in opium poppy (Desgagné-Penix and Facchini 2012). The VIGS-mediated suppression of CNMT transcript levels in opium poppy plants increased papaverine accumulation, whereas reduced transcript levels of N7OMT, which is specific to the N-desmethyl pathway, decreased papaverine levels. In contrast, suppression of 7OMT transcript levels did not affect papaverine accumulation in further support of the N-desmethyl pathway as the major route to papaverine. However, a minor contribution of the N-methyl pathway to papaverine biosynthesis cannot be ruled out.
Another study used a transcriptomics approach to compare high and low papaverine cultivars (Patahnk et al. 2013). Similarly, transcripts encoding enzymes in the N-desmethyl pathway were upregulated, whereas transcripts corresponding to 7OMT were downregulated.
The final step of papaverine biosynthesis involves oxidation of the fully O-methylated and N-desmethyl compound tetrahydropapaverine by dihydrobenzophenanthridine oxidase (DBOX), an enzyme that also catalyzes the final step in the formation of the quaternary benzo[c]phenanthridine sanguinarine (Hagel et al. 2012). Interestingly, DBOX transcripts occur exclusively in the roots of opium poppy suggesting the possible systemic translocation of papaverine from roots to aerial organs.
Morphine
Morphine biosynthesis has only been described in a few plant species restricted to the Papaveraceae, although the tetracyclic promorphinan alkaloid salutaridine and several derivatives have been reported in some members of the Euphorbiaceae (Theuns et al. 1986). The biosynthesis of morphine (Fig. 1) requires the epimerization of (S)-reticuline to (R)-reticuline. The reaction mechanism has been proposed to proceed via the dehydrogenation of (S)-reticuline to a 1,2-dehydroreticulinium ion (Hirata et al. 2004), which is subsequently reduced to (R)-reticuline. Although the 1,2-dehydroreticuline synthase (DRS) catalyzing the first conversion is not known, a 1,2-dehydroreticuline reductase (DRR) potentially involved in the second step has been purified to homogeneity and partially characterized (De-Eknamul and Zenk 1992). Although (S)-tetrahydroisoquinoline intermediates are assumed as the major route to (R,S)-reticuline, most of the enzymes involved accept both S and R epimers. In some plants, (R)-tetrahydroisoquinolines are required for the synthesis of BIA structural subgroups other than morphinan alkaloids, including the formation of the bisBIA berbamunine in Berberis stolonifera via the C–O coupling of (S)- and (R)-N-methylcoclaurine by the CYP berbamunine synthase (CYP80A1) (Fig. 1) (Stadler and Zenk 1993; Kraus and Kutchan 1995).
(R)-Reticuline is converted by the CYP salutaridine synthase (CYP719B1) to salutaridine (Gesell et al. 2009), which is then converted by the short-chain dehydrogenase/reductase salutaridine reductase (SalR) (Ziegler et al. 2006). Subsequently, salutaridinol is O-acetylated by salutaridinol 7-O-acetyltransferase (SalAT) (Grothe et al. 2001; Lenz and Zenk 1995). Interestingly, salutaridinol 7-O-acetate undergoes spontaneous cyclization to the first pentacyclic morphinan alkaloid thebaine at pH 8–9, whereas a dibenz[d,f]azonine alkaloid is formed at pH 6–7, suggesting compartmentalization of the intermediate in a basic environment or the involvement of an uncharacterized enzyme (Fisinger et al. 2007). A reduced derivative, neodihydrothebaine has only been reported in Papaver bracteatum (Theuns et al. 1984).
The morphine pathway bifurcates at thebaine with the initial step in the major route catalyzed by thebaine 6-O-demethylase (T6ODM) yielding neopinone, which undergoes purportedly spontaneous rearrangement to codeinone (Hagel and Facchini 2010). Codeinone is then reduced by the aldo–keto reductase codeinone reductase (COR) (Unterlinner et al. 1999) to codeine, which is converted to morphine by codeine O-demethylase (CODM). Alternatively, the CODM catalyzes the first step in the minor route, which involves the 3-O-demethylation of thebaine to oripavine. T6ODM then converts oripavine to morphinone, which is reduced by COR to morphine (Fig. 1).
The physiological role of BIA biosynthetic enzymes has been investigated using a variety of gene suppression and over-expression methods in opium poppy. The RNAi-mediated suppression of COR transcript levels resulted in the accumulation of reticuline and methylated 1-benzylisoquinoline derivatives with a corresponding decrease in morphinan alkaloid content (Allen et al. 2004). In support of a role for COR in the modulation of morphine biosynthesis, over-expression of COR resulted in a significant increase in morphinan alkaloid levels (Larkin et al. 2007). Similarly, over-expression of SalAT also increased morphinan alkaloid content, whereas suppression of SalAT transcript levels caused the accumulation of salutaridine, but did not alter the abundance or profile of morphinan alkaloids (Allen et al. 2008). The physiological roles of T6ODM and CODM were suggested in the opium poppy mutant top1 (Millgate et al. 2004), which accumulates high levels of thebaine and oripavine at the expense of codeine and morphine. The absence of T6ODM transcripts in a high-thebaine/oripavine, low-codeine/morphine opium poppy chemotype was compared to three high-codeine/morphine varieties to identify the corresponding gene using a microarray-based approach (Hagel and Facchini 2010). Suppression of T6ODM transcript levels resulted in the accumulation of thebaine and oripavine and substantially reduced levels of codeine and morphine. Similarly, suppression of CODM transcript levels caused the accumulation of codeine at the expense of morphine (Hagel and Facchini 2010). The isolation of all biosynthetic genes facilitated a coordinated confirmation of the physiological role of each enzyme operating downstream of (R)-reticuline (Wijekoon and Facchini 2012).
Protoberberines, protopines and sanguinarine
(S)-Scoulerine, the branch-point intermediate leading to protoberberine alkaloids, is formed from (S)-reticuline by BBE (Dittrich and Kutchan 1991; Facchini et al. 1996; Kutchan and Dittrich 1995; Winkler et al. 2006). (S)-Scoulerine is converted via methylenedioxy bridge formation and O-methylation to a variety of protoberberines including (S)-canadine, (S)-stylopine and (S)-sinactine. (S)-Canadine, the precursor to the protopine allocryptopine and the phthalideisoquinoline noscapine, is formed sequentially from (S)-scoulerine by (1) scoulerine 9-O-methyltransferase (SOMT1) (Dang and Facchini 2012; Takeshita et al. 1995) yielding tetrahydrocolumbamine and (2) the methylenedioxy bridge forming enzyme canadine synthase (CAS), which is a member of the CYP719A subfamily (Díaz Chávez et al. 2011; Ikezawa et al. 2003; Winzer et al. 2012). Similarly, (S)-sinactine is formed from (S)-scoulerine by (1) the addition of a methylenedioxy bridge by cheilanthifoline synthase, also a member of the CYP719A subfamily (Díaz Chávez et al. 2011; Ikezawa et al. 2009), followed by (2) 2-O-methylation of cheilanthifoline by an enzyme not yet characterized from opium poppy. A protoberberine 2-O-methyltransferase, columbamine O-methyltransferase, has been reported from meadow rue (Coptis japonica) (Morishige et al. 2002). Alternatively, cheilanthifoline can be oxidized to stylopine by another CYP719A, stylopine synthase (SPS) (Ikezawa et al. 2007; Díaz Chávez et al. 2011).
Protopine alkaloids are formed via the 14-hydroxylation of quaternary protoberberine alkaloids, which leads to ring tautomerization through C–N bond cleavage and formation of a C14 keto moiety. Quaternary protoberberines are formed by (S)-tetrahydroprotoberberine N-methyltransferase (TNMT) and 14-hydroxylation is catalyzed by (S)-cis-N-methylstylopine 14-hydroxylase (MSH). Opium poppy TNMT was shown to accept a variety of protoberberine substrates, but displays a preference for stylopine and canadine (Liscombe and Facchini 2007). MSH is a member of the CYP82 N subfamily and accepts a variety of quaternary protoberberines (Rueffer and Zenk 1987; Beaudoin and Facchini 2013). The protopines allocryptopine, cryptopine and protopine are formed from canadine, sinactine and stylopine, respectively. T6ODM, CODM and other ODDs are potentially involved in regulating protopine alkaloid biosynthesis through the oxidative dealkylation of methylenedioxy bridges or methoxy groups (Farrow and Facchini 2013).
Formation of the root-specific benzo[c]phenanthridine alkaloid dihydrosanguinarine (Fig. 1) from protopine is catalyzed by protopine 6-hydroxylase (P6H), another member of the CYP82 N subfamily (Tanahashi and Zenk 1990; Takemura et al. 2012). The resulting 6-hydroxylation results in a spontaneous rearrangement of 6-hydroxyprotopine to dihydrosanguinarine, which is oxidized to sanguinarine by the FAD-linked enzyme dihydrobenzophenanthridine oxidase (DBOX) (Hagel et al. 2012). Interestingly, sanguinarine can be reduced to dihydrosanguinarine by sanguinarine reductase (SanR) (Vogel et al. 2010; Weiss et al. 2006). The physiological role of SanR has been suggested as the mitigation of the cytotoxic effects of sanguinarine via conversion to the less-toxic dihydrosanguinarine.
Further oxidized protoberberines are common in some members of the Papaveraceae and are widespread in related plant families. In particular, berberine is formed from (S)-canadine by the FAD-linked enzyme (S)-tetrahydroprotoberberine oxidase (STOX) (Fig. 1) (Amann et al. 1984; Gesell et al. 2011; Matsushima et al. 2012). However, berberine and other modified protoberberines have generally not been reported in opium poppy (Shamma and Moniot 1978; Shulgin and Perry 2002). Moreover, protoberberines in which the berberine bridge has formed between the N-methyl and the 6′ carbons, as opposed to the 2′ position (i.e. yielding scoulerine), have been reported in other plants, but are not detected in opium poppy (Tsai and Lee 2010).
It is interesting to note that other species accumulate a variety of benzo[c]phenanthridine alkaloids derived from corresponding protoberberine alkaloids with differential methylation patterns. However, the only abundant benzo[c]phenanthridine alkaloids in opium poppy are dihydrosanguinarine and sanguinarine, which suggests that P6H might exhibit relatively narrow substrate specificity in opium poppy. Alternatively, the sanguinarine pathway could involve inter- or intra-cellular compartmentalization restricting the availability of substrates, other than protopine, to P6H.
Noscapine
In some opium poppy chemotypes, the phthalideisoquinoline alkaloid noscapine accounts for a majority of the total alkaloid content in latex (Frick et al. 2005). Noscapine also occurs in other members of the Papaveraceae and Menispermaceae. Owing to its cough-suppressing activity and potential as an anticancer drug (Mahmoudian and Rahimi-Moghaddam 2009), the biosynthesis of noscapine has been the subject of renewed interest (Dang and Facchini 2012; Facchini et al. 2007; Winzer et al. 2012). The formation of noscapine has not been as extensively investigated as other alkaloids, such as morphine. For decades, the role of scoulerine as a pathway intermediate was the extent of our knowledge concerning noscapine biosynthesis.
Based on the isolation of purported pathway intermediates and the identification of the enzymes involved in the formation of N-methylcanadine, a metabolic scheme for noscapine biosynthesis was proposed (Fig. 1); (Facchini et al. 2007). The scheme involves sequential hydroxylation of the isoquinoline ring, oxidative cleavage of the berberine bridge and two subsequent oxidations to form the characteristic phthalideisoquinoline lactone ring. Following the identification of relevant O-methyltransferases (Dang and Facchini 2012), a gene cluster proposed to encode all noscapine biosynthetic enzymes was identified in opium poppy. The functions for some gene cluster members were suggested using VIGS to show correlations between the suppression of a specific gene and the accumulation of compounds purported as pathway intermediates (Winzer et al. 2012). The function of metabolic gene clusters in plants has been suggested as a mechanism to regulate expression at the epigenetic level (Wegel et al. 2009) and to facilitate complete inheritance of a specialized metabolic pathway (Swaminathan et al. 2009). Recently, CAS (Dang and Facchini 2014a), N-methylcanadine 1-hydroxylase (CYP82Y1) (Dang and Facchini 2014b) and noscapine synthase (NOS) (Chen and Facchini 2014) were isolated and characterized from opium poppy. NOS was characterized as a NAD+-dependent, short-chain dehydrogenase/reductase catalyzing the irreversible conversion of narcotinehemiacetal to noscapine.
Other alkaloids
The rhoeadine alkaloids, some of which are also known as papaverrubine alkaloids, are found exclusively in members of the Papaveraceae, including opium poppy (Montgomery et al. 1983). Radiolabeling studies in Persian poppy (Papaver bracteatum) showed that the precursors to the rhoeadine alpinigenine were the protoberberines tetrahydropalmatine (Rönsch 1972), N-tetrahydropalamatine and the protopine muramine (Rönsch 1977). A route through protopines was confirmed in Papaver rhoeas by feeding protopine, which was converted to rhoeadine (Tani and Tagahara 1977). Labeling studies also showed that the pro-S-13-hydrogen of C13 is stereospecifically removed, suggesting that the cyclization is enzyme-mediated and not spontaneous (Battersby and Staunton 1974). Many rhoeadines isolated from opium poppy are 10-O-desmethylated (Montgomery et al. 1983). The VIGS-mediated suppression of gene transcripts encoding CODM, which catalyzes the O-demethylation and/or O,O-demethylenation of morphinan and protopine alkaloids, resulted in elevated levels of rhoeadine alkaloids, possibly owing to the increased accumulation of cryptopine and 10-O-desmethylcryptopine (Farrow and Facchini 2013). Enzymes involved in the conversion of protopines to rhoeadines are not known.
Regulation of alkaloid metabolism
Cellular localization and transport
The biosynthesis and storage of BIAs in opium poppy has been extensively investigated and shown to involve three distinct cell types (Facchini and De Luca 1995; Bird et al. 2003; Samanani et al. 2006; Lee and Facchini 2010). In situ hybridization and immunofluorescence labeling demonstrated the localization of biosynthetic gene transcripts and enzymes to companion cells and sieve elements, respectively. Since all characterized genes and enzymes involved in the formation of (S)-reticuline, and some functioning in various branch pathways, were initially localized to companion cells and sieve elements, respectively, laticifers were proposed to function only in the storage of alkaloids. The model proposed that BIA biosynthetic genes are expressed in companion cells and the corresponding enzymes are subsequently translocated to sieve elements. Alkaloids synthesized in sieve elements are finally transported to nearby laticifers for storage in large cytoplasmic vesicles.
However, SalAT and COR transcripts have also been reported in the cytoplasm, or latex, of laticifers (Decker et al. 2000; Larkin et al. 2007; Allen et al. 2008), although in situ hybridization and immunofluorescence labeling failed to detect SalAT and COR mRNA and corresponding enzymes in laticifers (Bird et al. 2003; Weid et al. 2004; Samanani et al. 2006). Recently, the application of shotgun proteomics to determine the relative abundance of proteins in isolated latex, or in whole stem including latex, revealed that the final three steps in morphine biosynthesis, catalyzed by T6ODM, COR and CODM, occur primarily in laticifers (Onoyovwe et al. 2013). Other BIA biosynthetic enzymes abundant in laticifers include NOS (Chen and Facchini 2014) and 7OMT (Onoyovwe et al. 2013), indicating that the final steps in the formation of noscapine and 7-O-methylated derivatives of reticuline also occur in latex and are spatially separated from upstream enzymes. Top candidates for transport from sieve elements to laticifers include thebaine and narcotinehemiacetal in the morphine and noscapine pathways, respectively. However, the detection of most pathway intermediates in isolated latex suggests a relatively non-specific transport process. Although the mechanism of translocation is unknown, the occurrence of plasmodesmata between opium poppy sieve elements and laticifers (Facchini and DeLuca 2008) suggests that both symplastic and apoplastic transport routes are possible. An ATP-binding cassette (ABC) transporter has been implicated in the uptake (Sakai et al. 2002) and efflux (Terasaka et al. 2003) of berberine in Coptis japonica, and the corresponding gene has been isolated (Shitan et al. 2003, 2012). However, it is not known whether an ABC transporter performs a similar function in opium poppy. The major sites of BIA biosynthetic gene transcript and enzyme localization and the possible points of translocation within the morphine pathway are summarized schematically in Fig. 2.
Cellular localization model showing the roles of companion cells, sieve elements and laticifers in the biosynthesis of morphine in opium poppy. Genes expressed in each cell type are shown in italics, whereas enzymes are indicated in red. The horizontal black arrow suggests thebaine as the major pathway intermediate translocated between sieve elements and laticifers. Font size is generally proportional to relative transcript and enzyme levels. Abbreviations are provided in the legend of Fig. 1
Subcellular compartmentalization and trafficking
In opium poppy, the antimicrobial alkaloid sanguinarine is not normally present in the latex, but accumulates constitutively in roots. The subcellular localization of most enzymes involved in the biosynthesis of sanguinarine has been investigated in cultured opium poppy cells using microprojectile bombardment followed by imaging of green fluorescent protein fusions (Hagel and Facchini 2012). As shown previously by density gradient fractionation (Amann et al. 1986; Rueffer and Zenk 1987; Tanahashi and Zenk 1990; Bauer and Zenk 1991), and using immunogold localization (Alcantara et al. 2005), BBE and CYPs associate with the endoplasmic reticulum (ER). Opium poppy BBE was also shown to contain a 25-amino acid, N-terminal signal peptide directing the enzyme to the ER, and an adjacent vacuolar-sorting determinant, resulting in its final translocation to the vacuole (Bird and Facchini 2001). Interestingly, the intracellular translocation of NCS was similar to that reported for BBE, predicting the extensive subcellular trafficking of sanguinarine biosynthetic pathway intermediates. The compartmentalization of NCS in the ER lumen requires the concomitant translocation of dopamine and 4HPAA, as depicted schematically in Fig. 3. As expected, O- and N-methyltransferases were detected as cytosolic enzymes; thus, the subsequent enzyme (6OMT) predicts the immediate export of (S)-norcoclaurine from the ER. The biosynthesis of (S)-reticuline could involve a metabolic channel anchored to the cytosolic face of the ER by NMCH, although no evidence is yet available to support interactions among the relevant enzymes. The co-compartmentalization of BBE in the ER lumen implies that (S)-reticuline is also imported from the cytosol for conversion to (S)-scoulerine, which is promptly exported. Four of the five subsequent enzymes are CYPs with active domains on the cytosolic face of the ER, with a cytosolic enzyme (TNMT) operating in the middle. Although not localized as a GFP fusion along with the other enzymes (Hagel and Facchini 2012), DBOX contains a predicted N-terminal signal peptide (Hagel et al. 2012) and related STOX enzymes have been associated with the ER (Amann et al. 1988); thus, a third reentry of a pathway intermediate into the ER lumen is predicted. Oxidation of dihydrosanguinarine in the ER would facilitate the vesicle-mediated transport of sanguinarine to the central vacuole of cultured opium poppy cells (Alcantara et al. 2005). The proposed subcellular trafficking features of sanguinarine biosynthesis might also occur in other BIA branch pathways, although no empirical evidence is available. Vacuolar transport in Coptis japonica has been reported to depend on a H+/berberine antiporter (Otani et al. 2005), but the process in opium poppy has not been investigated.
Subcellular localization model showing the proposed compartmentalization of sanguinarine biosynthetic enzymes in cultured opium poppy cells. The red arrow tracks the intracellular transport of pathway intermediates from the precursors dopamine and 4-hydroxyphenylacetaldehyde (4HPAA) to sanguinarine. The green arrow shows the proposed trafficking of sanguinarine from the lumen of the endoplasmic reticulum to the central vacuole. Abbreviations are provided in the legend of Fig. 1
Hormonal regulation
The phytohormonal activation of BIA metabolism has been used to identify biosynthetic genes in plants related to opium poppy. Addition of cytokinin to Thalictrum minus cell cultures caused a marked increase in the accumulation of berberine resulting from increased expression of 6OMT (Hara et al. 1994) and STOX (Hara et al. 1995). Ethylene was also shown to promote berberine biosynthesis in T. minus cell cultures, and the potent ethylene inhibitor silver thiosulfate caused a substantial decrease in berberine accumulation (Kobayashi et al. 1991).
Industry-sponsored research has focused on increasing the levels of morphinan pathway intermediates at the expense of morphine through the application of specific phytohormones or phytohormone catabolic inhibitors. The application of trinexapac-ethyl, an inhibitor of ODDs involved in gibberellin (GA) biosynthesis, with or without methyl jasmonate results in the introduction of a putative catabolic bottleneck leading to an increase in the relative abundance of thebaine and codeine and a decrease in the accumulation of morphine (Cotterill 2010). Interestingly, trinexapac-ethyl and other acylcyclohexanediones do not inhibit T6ODM and CODM directly (Hagel and Facchini 2010). Exogenous addition of GA3 was reported to increase the yield of morphine (Khan et al. 2007), suggesting a substantial role for phytohormone control in morphinan alkaloid metabolism. The developmental and phytohormonal control of BIA biosynthesis in opium poppy is a key area for future research, which could have direct consequences on the commercial production of opiate pharmaceuticals.
Transcriptional regulation
The transcriptional regulation of BIA biosynthesis has recently been investigated in opium poppy (Kawano et al. 2012; Mishra et al. 2013), but has been more extensively studied in Coptis japonica (Kato et al. 2007; Yamada et al. 2011). The importance of transcriptional control in BIA metabolism is particularly evident from (1) the cell type-specific localization of biosynthetic gene transcripts (Bird et al. 2003; Samanani et al. 2006; Lee and Facchini 2010; Onoyovwe et al. 2013) and (2) the coordinated induction of all expressed biosynthetic gene transcripts (Zulak et al. 2007) and corresponding proteins (Zulak et al. 2009; Desgagné-Penix et al. 2010) in opium poppy cell cultures after the addition of a fungal-derived elicitor.
The first transcriptional regulator of BIA metabolism was identified from C. japonica using RNAi to silence five putative regulatory factors showing reduced expression in a cultured cell line with relatively low BIA biosynthetic gene transcript levels (Kato et al. 2007). The transient silencing of one candidate (CjWRKY1) resulted in a marked decrease in berberine biosynthetic gene transcripts. In contrast, over-expression of CjWRKY1 increased berberine biosynthetic gene transcript levels. A bHLH transcriptional regulator involved in berberine biosynthesis was isolated using a similar approach (Yamada et al. 2011). In contrast, wounding and other stimuli to elicit a defense response was used to identify expressed sequence tags corresponding to induced gene transcripts in opium poppy plants (Mishra et al. 2013). A putative WKRY transcription factor was isolated and shown to bind conserved WRKY binding elements in promoters of certain biosynthetic genes in vitro and using a yeast one-hybrid approach. Another WRKY transcription factor was identified from opium poppy T-DNA insertion lines displaying altered BIA accumulation (Kawano et al. 2012). Moreover, the ectopic expression of an Arabidopsis thaliana WRKY transcription factor in California poppy (Eschscholzia californica) caused the induction of BBE and NMCH transcripts, and a substantial increase in the accumulation of benzo[c]phenanthridine alkaloids, including sanguinarine (Apuya et al. 2008). Interestingly, several putative WRKY elements occur within or near the promoter regions of biosynthetic genes in the reported noscapine gene cluster (Winzer et al. 2012), suggesting that noscapine biosynthesis might also be regulated by WRKY factors.
Metabolic engineering
Plants
Metabolic engineering has been used to modulate alkaloid composition and yield in opium poppy plants, with mixed results. Opium poppy plants engineered with constitutively expressed or antisense-suppressed NMCH showed substantial modulations in overall alkaloid content, but the BIA profile was not altered (Frick et al. 2007). In contrast, over-expression of COR (Larkin et al. 2007) and SalAT (Allen et al. 2008) specifically resulted in increased morphinan alkaloid accumulation. Surprisingly, the RNAi-mediated silencing of COR (Allen et al. 2004) did not result in the accumulation of codeine, but rather caused a substantial accumulation of (S)-reticuline and a concomitant decrease in the levels of morphine pathway intermediates downstream of salutaridine. The biochemical basis for the accumulation of an intermediate several enzymatic steps upstream of the silenced gene is not known, but suggestions include the possible occurrence of (1) feedback regulation and (2) metabolic channels (Allen et al. 2004). The occurrence of multienzyme complexes in morphine biosynthesis is supported by the suppression of SalAT in using RNAi (Allen et al. 2008). SalAT-silenced plants showed increased levels of salutaridine, which is not normally abundant in opium poppy. The accumulation of salutaridine was unexpected since the SalAT enzymatic substrate (salutaridinol) did not accumulate. Thus, salutaridine might be channeled to thebaine through an enzyme complex that includes SalR and SalAT. Finally, over-expression of A. thaliana transcriptional regulators in opium poppy resulted in increased thebaine and codeine accumulation associated with the up-regulation of several BIA biosynthetic enzymes (Apuya et al. 2008).
Microbes
Microbes engineered to express genes encoding BIA-biosynthetic enzymes provide a novel approach for the development of scalable manufacturing processes. The availability of large numbers of BIA biosynthetic genes from opium poppy and related plants has facilitated the reconstitution of several pathways leading to the production of (S)-reticuline and (R,S)-reticuline in E. coli (Minami et al. 2008) and Saccharomyces cerevisiae (Hawkins and Smolke 2008), respectively. Production of (S)-reticuline in E. coli was initially achieved by supplementing the bacterial culture medium with dopamine, some of which was converted to 3,4-dihydroxyphenylacetaldehyde (3,4-DHPAA) by a heterologously expressed bacterial monoamine oxidase (Minami et al. 2008). PR10-type NCS from C. japonica catalyzed the condensation of dopamine and 3,4-DHPAA to (S)-norlaudanosoline, which underwent successive methylations via 6OMT, CNMT and 4′OMT to yield (S)-reticuline. Recent modifications of this platform include the de novo synthesis of dopamine by the expression of two additional enzymes, tyrosinase and DOPA decarboxylase (Nakagawa et al. 2011). The use of bacterial enzymes facilitated the linking of BIA metabolism to the primary metabolism of E. coli, enabling a fermentation platform that creates plant products from simple carbon sources. In turn, E. coli-generated (S)-reticuline was converted to the aporphine alkaloid magnoflorine via co-culture with a strain of S. cerevisiae engineered to express the C. japonica genes encoding corytuberine synthase (CYP80G2) and an N-methyltransferase. Similarly, (S)-scoulerine was produced using a gene encoding BBE (Minami et al. 2008). Alternatively, (R,S)-reticuline was produced in S. cerevisiae from (R,S)-norlaudanosoline. The biosynthesis of both (R)- and (S)-reticuline isomers, coupled with enzymes derived from three different plant species and humans, was also used to produce protoberberine alkaloids and salutaridine (Hawkins and Smolke 2008).
Future prospects
Our knowledge of alkaloid biosynthesis in opium poppy has undergone two periods of major advancement: the first, several decades ago, involving elucidation of the chemistry of some pathways and the second, over the past decade, highlighting the isolation of most key biosynthetic genes largely facilitated by rapid developments in genomics. The availability of complete pathways at the genetic level will facilitate renewed advances in plant breeding and metabolic engineering and could lead to the establishment of alternative production systems, perhaps the most revolutionary being microbes. Beyond the isolation and characterization of enzymes, aspects of alkaloid biosynthesis that still require considerable research include (1) pathway regulation, at the levels of both gene expression and metabolic biochemistry, and (2) cellular biology, especially the inter- and intra-cellular transport processes involved in the formation and storage of compounds. The dedicated efforts of a relatively small number of research groups over the past half-century have generated an impressive body of information and elevated opium poppy to the status of a model system in the study of alkaloid biosynthesis in plants.
References
Alcantara J, Bird DA, Franceschi VR, Facchini PJ (2005) Sanguinarine biosynthesis is associated with the endoplasmic reticulum in cultured opium poppy cells after elicitor treatment. Plant Physiol 138:173–183. doi:10.1104/pp.105.059287
Allen RS, Millgate AG, Chitty JA, Thisleton J, Miller JAC, Fist AJ, Gerlach WL, Larkin PJ (2004) RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat Biotechnol 22:1559–1566. doi:10.1038/nbt1033
Allen RS, Miller JAC, Chitty JA, Fist AJ, Gerlach WL, Larkin PJ (2008) Metabolic engineering of morphinan alkaloids by over-expression and RNAi suppression of salutaridinol 7-O-acetyltransferase in opium poppy. Plant Biotechnol J 6:22–30. doi:10.1111/j.1467-7652.2007.00293.x
Amann M, Nagakura N, Zenk MH (1984) (S)-Tetrahydroprotoberberine oxidase the final enzyme in protoberberine biosynthesis. Tetrahedron Lett 25:953–954. doi:10.1016/S0040-4039(01)80071-x
Amann M, Wanner G, Zenk MH (1986) Intracellular compartmentation of two enzymes of berberine biosynthesis in plant cell cultures. Planta 167:310–320. doi:10.1007/BF00391333
Amann M, Nagakura N, Zenk MH (1988) Purification and properties of (S)-tetrahydroprotoberberine oxidase from suspension-cultured cells of Berberis wilsoniae. Eur J Biochem 175:17–25. doi:10.1111/j.1432-1033.1988.tb14160.x
Apuya NR, Park J-H, Zhang L, Ahyow M, Davidow P, Van Fleet J, Rarang JC, Hippley M, Johnson TW, Yoo H-D, Trieu A, Krueger S, Wu C, Lu Y, Flavell RB, Bobzin SC (2008) Enhancement of alkaloid production in opium and California poppy by transactivation using heterologous regulatory factors. Plant Biotechnol J 6:160–175. doi:10.1111/j.1467-7652.2007.00302.x
Battersby A, Staunton J (1974) Stereospecificity of some enzymic reactions. Tetrahedron 30:1707–1715. doi:10.1016/S0040-4020(01)97304-1
Bauer W, Zenk MH (1991) Two methylenedioxy bridge forming cytochrome P-450 dependent enzymes are involved in (S)-stylopine biosynthesis. Phytochemistry 30:2953–2962. doi:10.1016/S0031-9422(00)98230-X
Beaudoin GAW, Facchini PJ (2013) Isolation and characterization of a cDNA encoding (S)-cis-N-methylstylopine 14-hydroxylase from opium poppy, a key enzyme in sanguinarine biosynthesis. Biochem Biophys Res Comm 431:597–603. doi:10.1016/j.bbrc.2012.12.129
Berényi S, Csutorás C, Sipos A (2009) Recent developments in the chemistry of thebaine and its transformation products as pharmacological targets. Curr Med Chem 16:3215–3242. doi:10.2174/092986709788803295
Bird DA, Facchini PJ (2001) Berberine bridge enzyme, a key branch-point enzyme in benzylisoquinoline alkaloid biosynthesis, contains a vacuolar sorting determinant. Planta 213:888–897. doi:10.1007/s004250100582
Bird DA, Franceschi VR, Facchini PJ (2003) A tale of three cell types: alkaloid biosynthesis is localized to sieve elements in opium poppy. Plant Cell 15:2626–2635. doi:10.1105/tpc.015396
Brochmann-Hanssen E, Chen C, Chen CR, Chiang H, Leung AY, McMurtrey K (1975) Opium alkaloids. Part XVI. The biosynthesis of 1-benzylisoquinolines in P. somniferum. Preferred and secondary pathways; stereochemical aspects. J Chem Soc Perkin Trans 1:1531–1537. doi:10.1039/p19750001531
Chen X, Facchini PJ (2014) Short-chain dehydrogenase/reductase catalyzing the final step of noscapine biosynthesis is localized to laticifers in opium poppy. Plant J 77:173–184. doi:10.1111/tpj.12379
Choi K, Morishige T, Sato F (2001) Purification and characterization of coclaurine N-methyltransferase from cultured C. japonica cells. Phytochemistry 56:649–655. doi:10.1016/S0031-9422(00)00481-7
Cotterill P (2010) Method of altering the alkaloid composition in poppy plants. International Patent WO/2005/107436
Custers JHHV, Harrison SJ, Sela-Buurlage MB, van Deventer E, Lageweg W, Howe PW, van der Meijs PJ, Ponstein AS, Simons BH, Melchers LS, Stuiver MH (2004) Isolation and characterisation of a class of carbohydrate oxidases from higher plants, with a role in active defence. Plant J 39:147–160. doi:10.1111/j.1365-313X.2004.02117.x
Dang TTT, Facchini PJ (2012) Characterization of three O-methyltransferases involved in noscapine biosynthesis in opium poppy. Plant Physiol 159:618–631. doi:10.1104/pp.112.194886
Dang TTT, Facchini PJ (2014a) Cloning and characterization of canadine synthase involved in noscapine biosynthesis in opium poppy. FEBS Lett 588:198–204. doi:10.1016/j.febslet.2013.11.037
Dang TTT, Facchini PJ (2014b) CYP82Y1 is N-methylcanadine 1-hydroxylase, a key noscapine biosynthetic enzyme in opium poppy. J Biol Chem 289:2013–2026. doi:10.1074/jbc.M113.505099
Decker G, Wanner G, Zenk MH, Lottspeich F (2000) Characterization of proteins in latex of the opium poppy (P. somniferum) using two-dimensional gel electrophoresis and microsequencing. Electrophoresis 21:3500–3516. doi:10.1002/1522-2683(20001001)21:16<3500:AID-ELPS3500>3.0.CO;2-O
De-Eknamul W, Zenk MH (1992) Purification and properties of 1,2-dehydroreticuline reductase from P. somniferum seedlings. Phytochemistry 31:813–821. doi:10.1016/0031-9422(92)80020-F
Desgagné-Penix I, Facchini PJ (2012) Systematic silencing of benzylisoquinoline alkaloid biosynthetic genes reveals the major route to papaverine in opium poppy. Plant J 72:331–344. doi:10.1111/j.1365-313X.2012.05084.x
Desgagné-Penix I, Khan M, Schriemer DC, Cram D, Nowak J, Facchini PJ (2010) Integration of deep transcriptome and proteome analyses reveals the components of alkaloid metabolism in opium poppy cell cultures. BMC Plant Biol 10:252. doi:10.1186/1471-2229-10-252
Díaz Chávez ML, Rolf M, Gesell A, Kutchan TM (2011) Characterization of two methylenedioxy bridge-forming cytochrome CYP-dependent enzymes of alkaloid formation in the Mexican prickly poppy Argemone mexicana. Arch Biochem Biophys 507:186–193. doi:10.1016/j.abb.2010.11.016
Dittrich H, Kutchan TM (1991) Molecular cloning, expression, and induction of berberine bridge enzyme, an enzyme essential to the formation of benzophenanthridine alkaloids in the response of plants to pathogenic attack. Proc Natl Acad Sci USA 88:9969–9973. doi:10.1073/pnas.88.22.9969
Facchini PJ, De Luca V (1994) Differential and tissue-specific expression of a gene family for tyrosine/dopa decarboxylase in opium poppy. J Biol Chem 269:26684–26690
Facchini PJ, De Luca V (1995) Phloem-specific expression of tyrosine/DOPA decarboxylase genes and the biosynthesis of isoquinoline alkaloids in opium poppy. Plant Cell 7:1811–1821. doi:10.1105/tpc.7.11.1811
Facchini PJ, DeLuca V (2008) Opium poppy and Madagascar periwinkle: model non-model systems to investigate alkaloid biosynthesis in plants. Plant J 54:763–784. doi:10.1111/j.1365-313X.2008.03438.x
Facchini P, Penzes C, Johnson A, Bull D (1996) Molecular characterization of berberine bridge enzyme genes from opium poppy. Plant Physiol 112:1669–1677. doi:10.1104/pp.112.4.1669
Facchini PJ, Hagel JM, Liscombe DK, Loukanina N, MacLeod BP, Samanani N, Zulak KG (2007) Opium poppy: blueprint for an alkaloid factory. Phytochem Rev 6:97–124. doi:10.1007/s11101-006-9042-0
Farrow SC, Facchini PJ (2013) Dioxygenases catalyze O-demethylation and O,O-demethylenation with widespread roles in benzylisoquinoline alklaoid metabolism in opium poppy. J Biol Chem 288:28997–29012. doi:10.1016/j.phytochem.2012.02.014
Fisinger U, Grobe N, Zenk MH (2007) Thebaine synthase: a new enzyme in the morphine pathway in P. somniferum. Nat Prod Commun 2:249–253
Frick S, Kramell R, Schmidt J, Fist AJ, Kutchan TM (2005) Comparative qualitative and quantitative determination of alkaloids in narcotic and condiment P. somniferum cultivars. J Nat Prod 68:666–673. doi:10.1021/np0496643
Frick S, Kramell R, Kutchan TM (2007) Metabolic engineering with a morphine biosynthetic CYP in opium poppy surpasses breeding. Metabol Eng 9:169–176. doi:10.1016/j.ymben.2006.10.004
Gesell A, Rolf M, Ziegler J, Díaz Chávez ML, Huang F-C, Kutchan TM (2009) CYP719B1 is salutaridine synthase, the C–C phenol-coupling enzyme of morphine biosynthesis in opium poppy. J Biol Chem 284:24432–24442. doi:10.1074/jbc.M109.033373
Gesell A, Chávez MLD, Kramell R, Piotrowski M, Macheroux P, Kutchan TM (2011) Heterologous expression of two FAD-dependent oxidases with (S)-tetrahydroprotoberberine oxidase activity from A. mexicana and Berberis wilsoniae in insect cells. Planta 233:1185–1197. doi:10.1007/s00425-011-1357-4
Grothe T, Lenz R, Kutchan TM (2001) Molecular characterization of the salutaridinol 7-O-acetyltransferase involved in morphine biosynthesis in opium poppy P. somniferum. J Biol Chem 276:30717–30723. doi:10.1074/jbc.M102688200
Hagel JM, Facchini PJ (2010) Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat Chem Biol 6:273–275. doi:10.1038/nchembio.317
Hagel JM, Facchini PJ (2012) Subcellular localization of sanguinarine biosynthetic enzymes in cultured opium poppy cells. In Vitro Cell Dev Biol Plant 48:233–240. doi:10.1007/s11627-012-9426-3
Hagel JM, Facchini PJ (2013) Benzylisoquinoline alkaloid metabolism: a century of discovery and a brave new world. Plant Cell Physiol 54:647–672. doi:10.1093/pcp/pct020
Hagel JM, Beaudoin GAW, Fossati E, Ekins A, Martin VJJ, Facchini PJ (2012) Characterization of a flavoprotein oxidase from opium poppy catalyzing the final steps in sanguinarine and papaverine biosynthesis. J Biol Chem 287:42972–42983. doi:10.1074/jbc.M112.420414
Han X, Lamshöft M, Grobe N, Ren X, Fist AJ, Kutchan TM, Spiteller M, Zenk MH (2010) The biosynthesis of papaverine proceeds via (S)-reticuline. Phytochemistry 71:1305–1312. doi:10.1016/j.phytochem.2010.04.022
Hara M, Tanaka S, Tabata M (1994) Induction of a specific methyltransferase activity regulating berberine biosynthesis by cytokinin in T. minus cell cultures. Phytochemistry 36:327–332. doi:10.1016/S0031-9422(00)97070-5
Hara M, Morio K, Yazaki K, Tanaka S, Tabata M (1995) Separation and characterization of cytokinin-inducible (S)-tetrahydroberberine oxidases controlling berberine biosynthesis in T. minus cell cultures. Phytochemistry 38:89–93. doi:10.1016/0031-9422(94)00623-2
Hawkins KM, Smolke CD (2008) Production of benzylisoquinoline alkaloids in S. cerevisiae. Nat Chem Biol 4:564–573. doi:10.1038/nchembio.105
Hirata K, Poeaknapo C, Schmidt J, Zenk MH (2004) 1,2-Dehydroreticuline synthase, the branch point enzyme opening the morphinan biosynthetic pathway. Phytochemistry 65:1039–1046. doi:10.1016/j.phytochem.2004.02.015
Ibrahim RK, Muzac I (2000) The methyltransferase gene superfamily: a tree with multiple branches. Recent Adv Phytochemistry 34:349–384. doi:10.1016/S0079-9920(00)80012-X
Ikezawa N, Tanaka M, Nagayoshi M, Shinkyo R, Sakaki T, Inouye K, Sato F (2003) Molecular cloning and characterization of CYP719, a methylenedioxy bridge-forming enzyme that belongs to a novel CYP family, from cultured C. japonica cells. J Biol Chem 278:38557–38565. doi:10.1074/jbc.M302470200
Ikezawa N, Iwasa K, Sato F (2007) Molecular cloning and characterization of methylenedioxy bridge-forming enzymes involved in stylopine biosynthesis in Eschscholzia californica. FEBS J 274:1019–1035. doi:10.1111/j.1742-4658.2007.05652.x
Ikezawa N, Iwasa K, Sato F (2009) CYP719A subfamily of cytochrome CYP oxygenases and isoquinoline alkaloid biosynthesis in E. californica. Plant Cell Rep 28:123–133. doi:10.1007/s00299-008-0624-8
Kato N, Dubouzet E, Kokabu Y, Yoshida S, Taniguchi Y, Dubouzet JG, Yazaki K, Sato F (2007) Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in C. japonica. Plant Cell Physiol 48:8–18. doi:10.1093/pcp/pcl041
Kawano N, Kiuchi F, Kawahara N, Yoshimatsu K (2012) Genetic and phenotypic analyses of a P. somniferum T-DNA insertional mutant with altered alkaloid composition. Pharmaceuticals 5:133–154. doi:10.3390/ph5020133
Khan R, Khan MMA, Singh M, Nasir S, Naeem M, Siddiqui MH, Mohammad F (2007) Gibberellic acid and triacontanol can ameliorate the opium yield and morphine production in opium poppy (P. somniferum L.). Acta Agric Scandinavica B 57:307–312. doi:10.1080/09064710600982811
Kobayashi Y, Hara M, Fukui H, Tabata M (1991) The role of ethylene in berberine production by T. minus cell suspension cultures. Phytochemistry 30:3605–3609. doi:10.1016/0031-9422(91)80075-C
Kraus PF, Kutchan TM (1995) Molecular cloning and heterologous expression of a cDNA encoding berbamunine synthase, a C–O phenol-coupling cytochrome P450 from the higher plant Berberis stolonifera. Proc Natl Acad Sci USA 92:2071–2075
Kutchan TM, Dittrich H (1995) Characterization and mechanism of the berberine bridge enzyme, a covalently flavinylated oxidase of benzophenanthridine alkaloid biosynthesis in plants. J Biol Chem 270:24475–24481. doi:10.1074/jbc.270.41.24475
Larkin PJ, Miller JAC, Allen RS, Chitty JA, Gerlach WL, Frick S, Kutchan TM, Fist AJ (2007) Increasing morphinan alkaloid production by over-expressing codeinone reductase in transgenic P. somniferum. Plant Biotechnol J 5:26–37. doi:10.1111/j.1467-7652.2006.00212.x
Lee E-J, Facchini P (2010) Norcoclaurine synthase is a member of the pathogenesis-related 10/Bet v1 protein family. Plant Cell 22:3489–3503. doi:10.1105/tpc.110.077958
Lee E-J, Facchini PJ (2011) Tyrosine aminotransferase contributes to benzylisoquinoline alkaloid biosynthesis in opium poppy. Plant Physiol 157:1067–1078. doi:10.1104/pp.111.185512
Lenz R, Zenk MH (1995) Acetyl coenzyme A:salutaridinol-7-O-acetyltransferase from P. somniferum plant cell cultures. The enzyme catalyzing the formation of thebaine in morphine biosynthesis. J Biol Chem 270:31091–31096. doi:10.1074/jbc.270.52.31091
Liscombe DK, Facchini PJ (2007) Molecular cloning and characterization of tetrahydroprotoberberine cis-N-methyltransferase, an enzyme involved in alkaloid biosynthesis in opium poppy. J Biol Chem 282:14741–14751. doi:10.1074/jbc.M611908200
Liscombe DK, Macleod BP, Loukanina N, Nandi OI, Facchini PJ (2005) Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry 66:1374–1393. doi:10.1016/j.phytochem.2005.04.029
Liscombe DK, Ziegler J, Schmidt J, Ammer C, Facchini PJ (2009) Targeted metabolite and transcript profiling for elucidating enzyme function: isolation of novel N-methyltransferases from three benzylisoquinoline alkaloid-producing species. Plant J 60:729–743. doi:10.1111/j.1365-313X.2009.03980.x
Mahmoudian M, Rahimi-Moghaddam P (2009) The anti-cancer activity of noscapine: a review. Recent Pat Anticancer Drug Discov 4:92–97. doi:10.2174/157489209787002524
Matsushima Y, Minami H, Hori K, Sato F (2012) Pathway engineering of benzylisoquinoline alkaloid biosynthesis in transgenic California poppy cells with ectopic expression of tetrahydroberberine oxidase from C. japonica. Plant Biotechnol 29:473–481. doi:10.5511/plantbiotechnology.12.1101a
Millgate AG, Pogson BJ, Wilson IW, Kutchan TM, Zenk MH, Gerlach WL, Fist AJ, Larkin PJ (2004) Analgesia: morphine-pathway block in top1 poppies. Nature 431:413–414. doi:10.1038/431413a
Minami H, Dubouzet E, Iwasa K, Sato F (2007) Functional analysis of norcoclaurine synthase in C. japonica. J Biol Chem 282:6274–6282. doi:10.1074/jbc.M608933200
Minami H, Kim J-S, Ikezawa N, Takemura T, Katayama T, Kumagai H, Sato F (2008) Microbial production of plant benzylisoquinoline alkaloids. Proc Natl Acad Sci USA 105:7393–7398. doi:10.1073/pnas.0802981105
Mishra S, Triptahi V, Singh S et al (2013) Wound induced transcriptional regulation of benzylisoquinoline pathway and characterization of wound inducible PsWRKY transcription factor from P. somniferum. PLoS One 8:e52784. doi:10.1371/journal.pone.0052784
Montgomery CT, Cassels BK, Shamma M (1983) The rhoeadine alkaloids. J Nat Prod 46:441–453. doi:10.1021/np50028a001
Morishige T, Tsujita T, Yamada Y, Sato F (2000) Molecular characterization of the S-adenosyl-l-methionine:3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in C. japonica. J Biol Chem 275:23398–23405. doi:10.1074/jbc.M002439200
Morishige T, Dubouzet E, Choi K-B, Yazaki K, Sato F (2002) Molecular cloning of columbamine O-methyltransferase from cultured C. japonica cells. Eur J Biochem 269:5659–5667. doi:10.1046/j.1432-1033.2002.03275.x
Nakagawa A, Minami H, Kim J-S, Koyanagi T, Katayama T, Sato F, Kumagai H (2011) A bacterial platform for fermentative production of plant alkaloids. Nat Comm 2:326. doi:10.1038/ncomms1327
Onoyovwe A, Hagel JM, Chen X, Khan MF, Schriemer D, Facchini PJ (2013) Morphine biosynthesis in opium poppy involves two cell types: sieve elements and laticifers. Plant Cell 25:4110–4122. doi:10.1105/tpc.113.115113
Otani M, Shitan N, Sakai K, Martinoia E, Sato F, Yazaki K (2005) Characterization of vacuolar transport of the endogenous alkaloid berberine in C. japonica. Plant Physiol 138:1939–1946. doi:10.1104/pp.105.064352
Ounaroon A, Decker G, Schmidt J, Lottspeich F, Kutchan TM (2003) (R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum––cDNA cloning and characterization of methyl transfer enzymes of alkaloid biosynthesis in opium poppy. Plant J 36:808–819. doi:10.1046/j.1365-313X.2003.01928.x
Patahnk S, Deepika L, Gupta P, Mishra BK, Shukla S, Asif MH, Trivedi PK (2013) Comparative transcriptome analysis using high papaverine mutant of P. somniferum reveals pathway and uncharacterized steps of papaverine biosynthesis. PLoS One 8:e65622. doi:10.1371/journal.pone.0065622
Pauli HH, Kutchan TM (1998) Molecular cloning and functional heterologous expression of two alleles encoding (S)-N-methylcoclaurine 3′-hydroxylase (CYP80B1), a new methyl jasmonate-inducible cytochrome P-450-dependent mono-oxygenase of benzylisoquinoline alkaloid biosynthesis. Plant J 13:793–801. doi:10.1046/j.1365-313X.1998.00085.x
Pienkny S, Brandt W, Schmidt J, Kramell R, Ziegler J (2009) Functional characterization of a novel benzylisoquinoline O-methyltransferase suggests its involvement in papaverine biosynthesis in opium poppy (P. somniferum L). Plant J 60:56–67. doi:10.1111/j.1365-313X.2009.03937.x
Prescott AG, John P (1996) Dioxygenases: molecular structure and role in plant metabolism. Annu Rev Plant Physiol Plant Mol Biol 47:245–271. doi:10.1146/annurev.arplant.47.1.245
Rönsch H (1972) Zur Biosynthese der Alkaloide vom Rhoeadin-Typ. Bildung von Alpinigenin aus einer Tetrahydroprotoberberin-Vorstufe und Tetrahydropalmatin-methosalz. Eur J Biochem 28:123–126. doi:10.1111/j.1432-1033.1972.tb01892.x
Rönsch H (1977) Biosynthesis of alpinigenine by way of tetrahydroprotoberberine and protopine intermediates. Phytochemistry 16:691–698. doi:10.1016/S0031-9422(00)89233-X
Rueffer M, Zenk MH (1987) Enzymatic formation of protopines by a microsomal cytochrome P-450 system of Corydalis vaginans. Tetrahedron Lett 28:5307–5310. doi:10.1016/S0040-4039(00)96715-7
Sakai K, Shitan N, Sato F, Ueda K, Yazaki K (2002) Characterization of berberine transport into C. japonica cells and the involvement of ABC protein. J Exp Bot 53:1879–1886. doi:10.1093/jxb/erf052
Samanani N, Liscombe DK, Facchini PJ (2004) Molecular cloning and characterization of norcoclaurine synthase, an enzyme catalyzing the first committed step in benzylisoquinoline alkaloid biosynthesis. Plant J 40:302–313. doi:10.1111/j.1365-313X.2004.02210.x
Samanani N, Alcantara J, Bourgault R, Zulak KG, Facchini PJ (2006) The role of phloem sieve elements and laticifers in the biosynthesis and accumulation of alkaloids in opium poppy. Plant J 47:547–563. doi:10.1111/j.1365-313X.2006.02801.x
Shamma M, Moniot JL (1978) Isoquinoline alkaloids research 1972–1977. Plenum, New York. doi:10.1007/978-1-4615-8819-1
Shitan N, Bazin I, Dan K, Obata K, Kigawa K, Ueda K, Sato F, Forestier C, Yazaki K (2003) Involvement of CjMDR1, a plant multidrug-resistance-type ATP-binding cassette protein, in alkaloid transport in C. japonica. Proc Natl Acad Sci USA 100:751–756. doi:10.1073/pnas.0134257100
Shitan N, Dalmas F, Dan K, Obata K, Kigawa K, Ueda K, Sato F, Forestier C, Yazaki K (2012) Characterization of C. japonica CjABCB2, an ATP-binding cassette protein involved in alkaloid transport. Phytochemistry 91:109–116. doi:10.1016/j.phytochem.2012.02.012
Shulgin AT, Perry WE (2002) The simple plant isoquinolines. Transform Press, Berkley
Sirikantaramas S, Morimoto S, Shoyama Y, Ishikawa Y, Wada Y, Shoyama Y, Taura F (2004) The gene controlling marijuana psychoactivity: molecular cloning and heterologous expression of Δ1-tetrahydrocannabinolic acid synthase from Cannabis sativa L. J Biol Chem 279:39767–39774. doi:10.1074/jbc.M403693200
Stadler R, Zenk MH (1993) The purification and characterization of a unique cytochrome P-450 enzyme from Berberis stolonifera plant cell cultures. J Biol Chem 268:823–831
Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ (2009) CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21:3315–3325. doi:10.1105/tpc.108.063677
Takemura T, Ikezawa N, Iwasa K, Sato F (2012) Molecular cloning and characterization of a cytochrome CYP in sanguinarine biosynthesis from E. californica cells. Phytochemistry 91:100–108. doi:10.1016/j.phytochem.2012.02.013
Takeshita N, Fujiwara H, Mimura H, Fitchen JH, Yamada Y, Sato F (1995) Molecular cloning and characterization of S-adenosyl-l-methionine:scoulerine-9-O-methyltransferase from cultured cells of C. japonica. Plant Cell Physiol 36:29–36
Tanahashi T, Zenk MH (1990) Elicitor induction and characterization of microsomal protopine-6-hydroxylase, the central enzyme in benzophenanthridine alkaloid biosynthesis. Phytochemistry 29:1113–1122. doi:10.1016/0031-9422(90)85414-B
Tani C, Tagahara K (1977) Studies on the alkaloids of the papaveraceous plants. XXVIII. The biosynthesis of rhoeadine. J Pharm Soc Jpn 97:93–102
Terasaka K, Sakai K, Sato F, Yamamoto H, Yazaki K (2003) Thalictrum minus cell cultures and ABC-like transporter. Phytochemistry 62:483–489
Theuns HG, Lenting HBM, Salemink CA, Tanaka H, Shibata M, Ito K, J.J. Ch Lousberg R (1984) Neodihydrothebaine and bractazonine, two dibenz[d, f]azonine alkaloids of P. bracteatum. Phytochemistry 23:1157–1166. doi:10.1016/S0031-9422(00)82630-8
Theuns HG, Theuns HL, Lousberg RJJC (1986) Search for new natural sources of morphinans. Econ Bot 40:485–497. doi:10.1007/BF02859662
Tsai S-F, Lee S-S (2010) Characterization of acetylcholinesterase inhibitory constituents from Annona glabra assisted by HPLC microfractionation. J Nat Prod 73:1632–1635. doi:10.1021/np100247r
Unterlinner B, Lenz R, Kutchan TM (1999) Molecular cloning and functional expression of codeinone reductase: the penultimate enzyme in morphine biosynthesis in the opium poppy P. somniferum. Plant J 18:465–745. doi:10.1016/0031-9422(75)85346-5
Uprety H, Bhakuni DS, Kapil RS (1975) Biosynthesis of papaverine. Phytochemistry 14:1535–1537. doi:10.1016/0031-9422(75)85346-5
Vogel M, Lawson M, Sippl W, Conrad U, Roos W (2010) Structure and mechanism of sanguinarine reductase, an enzyme of alkaloid detoxification. J Biol Chem 285:18397–18406. doi:10.1074/jbc.M109.088989
Wegel E, Koumproglou R, Shaw P, Osbourn A (2009) Cell type-specific chromatin decondensation of a metabolic gene cluster in oats. Plant Cell 21:3926–3936. doi:10.1105/tpc.109.072124
Weid M, Ziegler J, Kutchan TM (2004) The roles of latex and the vascular bundle in morphine biosynthesis in the opium poppy, P. somniferum. Proc Natl Acad Sci USA 101:13957–13962. doi:10.1073/pnas.0405704101
Weiss D, Baumert A, Vogel M, Roos W (2006) Sanguinarine reductase, a key enzyme of benzophenanthridine detoxification. Plant Cell Environ 29:291–302. doi:10.1111/j.1365-3040.2005.01421.x
Wijekoon CP, Facchini PJ (2012) Systematic knockdown of morphine pathway enzymes in opium poppy using virus-induced gene silencing. Plant J 69:1052–1063. doi:10.1111/j.1365-313X.2011.04855.x
Winkler A, Hartner F, Kutchan TM, Glieder A, Macheroux P (2006) Biochemical evidence that berberine bridge enzyme belongs to a novel family of flavoproteins containing a bi-covalently attached FAD cofactor. J Biol Chem 281:21276–21285. doi:10.1074/jbc.M603267200
Winzer T, Gazda V, He Z, Kaminski F, Kern M, Larson TR, Li Y, Meade F, Teodor R, Vaistij FE, Walker C, Bowser TA, Graham IA (2012) A P. somniferum ten-gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336:1704–1708. doi:10.1126/science.1220757
Yamada Y, Kokabu Y, Chaki K, Yoshimoto T, Ohgaki M, Yoshida S, Kato N, Koyama T, Sato F (2011) Isoquinoline alkaloid biosynthesis is regulated by a unique bHLH-type transcription factor in C. japonica. Plant Cell Physiol 52:1131–1141. doi:10.1093/pcp/pcr062
Ziegler J, Voigtländer S, Schmidt J, Kramell R, Miersch O, Ammer C, Gesell A, Kutchan TM (2006) Comparative transcript and alkaloid profiling in Papaver species identifies a short chain dehydrogenase/reductase involved in morphine biosynthesis. Plant J 48:177–192. doi:10.1111/j.1365-313X.2006.02860.x
Zulak KG, Cornish A, Daskalchuk TE, Deyholos MK, Goodenowe DB, Gordon PMK, Klassen D, Pelcher LE, Sensen CW, Facchini PJ (2007) Gene transcript and metabolite profiling of elicitor-induced opium poppy cell cultures reveals the coordinate regulation of primary and secondary metabolism. Planta 225:1085–1106. doi:10.1007/s00425-006-0419-5
Zulak KG, Khan MF, Alcantara J, Schriemer DC, Facchini PJ (2009) Plant defense responses in opium poppy cell cultures revealed by liquid chromatography–tandem mass spectrometry proteomics. Mol Cell Proteomics 8:86–98. doi:10.1074/mcp.M800211-MCP200
Acknowledgments
Research in the laboratory of the corresponding author was supported through funding from Genome Canada, Genome Alberta, the Government of Alberta, the Canada Foundation for Innovation Leaders Opportunity Fund, and the Natural Sciences and Engineering Research Council of Canada. P.J.F. holds the Canada Research Chair in Plant Metabolic Processes Biotechnology. G.A.W.B. is the recipient of scholarships from the Natural Sciences and Engineering Research Council of Canada, Alberta Innovates Technology Futures, and the fonds québécois de la recherche sur la nature et les technologies.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Beaudoin, G.A.W., Facchini, P.J. Benzylisoquinoline alkaloid biosynthesis in opium poppy. Planta 240, 19–32 (2014). https://doi.org/10.1007/s00425-014-2056-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-014-2056-8