Abstract
Hairy stripes in Tribolium are generated during blastoderm and germ band extension, but a direct role for Tc-h in trunk segmentation was not found. We have studied here several aspects of hairy function and expression in Tribolium, to further elucidate its role. First, we show that there is no functional redundancy with other hairy paralogues in Tribolium. Second, we cloned the hairy orthologue from Tribolium confusum and show that its expression mimics that of Tribolium castaneum, implying that stripe expression should be functional in some way. Third, we show that the dynamics of stripe formation in the growth zone is not compatible with an oscillatory mechanism comparable to the one driving the expression of hairy homologues in vertebrates. Fourth, we use parental RNAi experiments to study Tc-h function and we find that mandible and labium are particularly sensitive to loss of Tc-h, reminiscent of a pair-rule function in the head region. In addition, lack of Tc-h leads to cell death in the gnathal region at later embryonic stages, resulting in a detachment of the head. Cell death patterns are also altered in the midline. Finally, we have analysed the effect of Tc-h knockdown on two of the target genes of hairy in Drosophila, namely fushi tarazu and paired. We find that the trunk expression of Tc-h is required to regulate Tc-ftz, although Tc-ftz is itself also not required for trunk segmentation in Tribolium. Our results imply that there is considerable divergence in hairy function between Tribolium and Drosophila.
Similar content being viewed by others
Introduction
Drosophila hairy (h) forms part of the segmentation cascade at the level of the pair-rule genes (Nüsslein-Volhard and Wieschaus 1980; Ingham 1988). Mutations of h result in the deletion of the posterior part of every odd-numbered segment in the resulting larvae, thus reflecting a classical pair-rule phenotype (Jürgens et al. 1984 ). h negatively regulates the spatial expression of the pair-rule genes runt (Klingler and Gergen 1993), fushi tarazu (ftz; Carroll et al. 1988; Rushlow et al. 1989; Tsai and Gergen 1995), and paired (prd; Baumgartner and Noll 1990; Gutjahr et al. 1993), and was therefore classified as primary pair-rule gene.
Hairy homologues occur also in vertebrates (called her or hes genes) where some of them are involved in the generation of the somites. The somites can be envisaged as the vertebrate analogs to the segments in insects, although segmentation does not proceed in the ectoderm as in insects, but in the mesoderm (Tautz 2004). Analysis of the process of somite formation revealed a fundamentally different regulatory mode for the vertebrate hairy homologues during the generation of these segmental units. Reflecting segmentation in a cellular environment, the process is based on cell signaling factors of the Notch/Delta and other signaling pathways. These regulate oscillating waves of expression of several genes across the growth zone, including some of the her or hes genes (reviewed in Rida et al. 2004; Giudicelli and Lewis 2004).
Intriguingly, a hairy homologue is also expressed during segmentation in the spider Cupiennius salei and functional analysis of the Notch/Delta pathway by RNAi shows strong disruption of segment formation (Stollewerk et al. 2003). This raises the possibility that oscillating expression of hairy homologues may be an ancestral feature of bilateria, and that the Drosophila pattern of direct regulation through transcription factor gradients is highly derived (Tautz 2004). It is therefore of particular interest to study hairy function in short-germband insect embryos, which also undergo a cellular rather than blastodermal mode of segmentation.
Tribolium embryogenesis can be considered as a typical representative of short germ embryogenesis. Tc-h is expressed in pair-rule stripes during blastoderm stage and germband extension (Sommer and Tautz 1993), which would suggest a pair-rule function. However, Choe et al. (2006) found in their functional studies of the pair-rule gene homologues in Tribolium no indication for a direct involvement of Tc-h in trunk segmentation, although they describe some phenotypic effects on head development. Their results have suggested that the pair-rule gene function in Tribolium differs is in many ways from Drosophila. They found that the homologues of even skipped, runt, and odd skipped form a regulatory circuit, regulating each other, as well as target genes, during the extension of the germband. These findings imply that the pair-rule machinery was subject to significant changes during insect evolution.
We have studied here the function of Tc-h in detail. We clarified whether we are indeed dealing with the right homologue and whether its expression pattern is reasonably conserved between more closely related species. We were further interested to study whether the emergence of Tc-h stripes during germband elongation might reflect oscillatory waves of expression. Finally, we wanted to understand its function during segmentation of the head region, as well as its interaction with other pair-rule genes.
Material and methods
Beetle handling and stock keeping
Beetle stocks were essentially kept as described by Berghammer et al. (1999). All experiments were performed using the T. castaneum wild type strain “San Bernadino” and a wild type strain of T. confusum, provided by Dick Beeman, Kansas State University, if not indicated otherwise. Flour was kept at 65°C overnight to prevent parasitic infections.
Cloning of Tribolium confusum hairy homologue
The hairy homologue from T. confusum was cloned by screening a genomic library cloned in Lambda Fix II provided by Sue Brown (Kansas). As probe we used a 383 bp subcloned PCR fragment obtained from T. confusum genomic DNA as template with the primers 5′AAYAARCCNATHATGGARAAR 3′ and 5′YTGNAGRTGYTTNACNGTCAT3′ covering the HLH region. The insert of a positive clone was subcloned as a NotI fragment into a plasmid vector and fully sequenced (Acc. No. EU819553).
Embryo collection and fixation
Eggs were collected from 0–48 h at 30°C to gather all developmental stages before dorsal closure. The embryos were rinsed with tapwater, mildly dechorionated for 1.5 min in 50% bleach and rinsed with tapwater afterwards to remove residual bleach. Fixation was performed in scintillation vials containing 3 ml PEMS (0.1 M Pipes, 2 mM MgSO4, 1 mM EDTA, pH 6.9), 6 ml heptane, and 4% formaldehyde on a shaking platform for 25 min. The water phase was then substituted for 8 ml methanol and the vial vigorously shaken for 30 s, resulting in devitellinization of the embryos by methanol shock. Undevitellinized embryos were mechanically devitellinized by squeezing them through a syringe using a 19 G needle. Embryos were kept at −20°C in methanol for subsequent analysis.
Parental RNAi
Parental RNAi experiments were performed according to Bucher et al. (2004) with slight modifications. Approximatly 200 female pupae were fixed to microscope slides using double-sided tape (Scotch 665). Pupae were taken off the slides after injection and transferred to “culture vials” containing full grain flour in order to facilitate eclosion. The first eggs were collected approximately 5 days after injection and incubated at 33°C for 4 days to allow full development in order to assess the amount and strength of phenocopies. Eggs were collected every 48 h and fixed for subsequent analysis by in situ hybridization. Once a week, a 24-h collection was allowed to fully develop and cuticle preparations were performed in Hoyer’s medium according to standard procedures (Berghammer et al. 1999) in order to monitor the phenotype/phenocopies over time. Double-stranded RNA was synthesized from PCR templates using the T7 MEGAscript RNAi Kit (Ambion) without additional annealing steps and injected at a concentration of 2 μg/μl in H2O with 10% Phenol red. The injection solution was thoroughly centrifuged at 13,000×g before injection to pellet any particles and reduce clogging of the needle.
In situ hybridization and antibody staining
Whole mount in situ hybridizations were performed according to standard protocols (Tautz and Pfeifle 1989; Klingler and Gergen 1993). Immunological staining was performed as described by MacDonald and Struhl (1986) with slight modifications. For the analysis of LacZ protein distribution in the transgenic lines, an additional signal amplification step using a secondary biotinylated antibody and the Vectastatin ABC HRP KIT (Vector Labs) was introduced to the protocol. For apoptosis detection using the anti cleaved caspase3 antibody (Cell Signalling Technology, Inc.), amplifications steps were omitted and staining was performed using a secondary alkaline phosphatase coupled antibody.
Results
Analysis of hairy paralogues
The first Tribolium hairy homologue isolated showed high similarity to Drosophila hairy in terms of amino acid sequence as well as expression pattern (Sommer and Tautz 1993). However, five additional homologues with modest to high similarity are found in the full genome sequence of Tribolium (Tribolium Genome Sequencing Consortium 2008), raising the possibility that there might be a duplicated gene that substitutes at least for part of the hairy function in Tribolium. Phylogenetic comparison of all five sequences with Drosophila and mouse homologues does not support such a notion. The hairy gene that was identified first (acc. no. NP_001107765) shares the highest similarity with Drosophila hairy (Fig. 1). The closest other homologue (acc. no. XP_967694) is the apparent orthologue of Drosophila deadpan (Fig. 1). This is confirmed by in situ hybridization. Similar to the Drosophila deadpan gene (Bier et al. 1992), Tc-dpn is also expressed during segmentation and in a pan-neuronal pattern during neurogenesis (supplementary Fig. S1). The segmental stripes appear to arise in a similar way as the Tc-h stripes, but are much thinner (supplementary Fig. S1). Despite its double-segmental expression, Drosophila deadpan function does not overlap with the segmentation function of hairy and we find the same for Tribolium (see below).
Phylogenetic comparison of hairy-like sequences. Neighbor-joining distance tree using full amino acid sequences of all genes with an HLH domain and a C-terminal WRPW motif from Drosophila (Dm-), Tribolium (Tc-) and mouse (Mm-). The hey gene homologues from these species serve as outgroup. Bootstrap values higher than 60% (out of 1,000 runs) are shown at the branches. Accession numbers are added to the gene names where these are not unequivocal
Comparison to Tribolium confusum
To assess whether the appearance of the Tc-h stripes is conserved in a distantly related Tribolium species, we have cloned the orthologue of Tc-h from Tribolium confusum. It is known that the expression characteristics of her genes can differ substantially between fish species, i.e., these patterns can be subject to regulatory changes (Gajewski et al. 2006).
The overall alignment between the T. castaneum and T. confusum sequences shows a good conservation of the exons and a generally high divergence in non-coding regions, but with conserved blocks that may be related to functional elements (supplementary Fig. S2). This conservation–divergence pattern is comparable to the average patterns of gene comparisons between D. melanogaster and D. virilis, or zebrafish and Medaka and indicates, therefore, a similar evolutionary distance between T. confusum and T. castaneum.
The expression of the hairy orthologue in T. confusum is highly comparable to the one in T. castaneum (supplementary Fig. S3). However, the stripe formation in the growth zone appears to be more distinct, i.e., the inter-stripe clearing occurs faster, at least at the early stages (Fig. 2). Apart of this, all other relevant features of hairy expression are conserved, including the longer expression in stripe #1 (Fig. 2c,g) and the expression in midline cells (Fig. 2d,h).
The dynamics of stripe formation
In vertebrates, the stripe formation in the growth zone occurs in cyclic expression waves, emanating from the posterior end. This implies that a given cell goes through several on and off states before it becomes differentiated. Delta–Notch signaling is a major driver of this cyclic expression and this was also found for basal arthropods, such as spiders and millipedes. In Tribolium, we have so far not found any evidence for an involvement of Delta–Notch signaling in segmentation (Aranda 2006; supplementary Fig. S4). However, this would not rule out that cyclic expression occurs, since other signaling mechanisms might substitute Delta–Notch signaling in Tribolium.
Inspection of the expression pattern does not seem to provide clear evidence for an expression wave of hairy across the growth zone. Rather, it appears that one stripe arises after the other, with a more or less fast clearance of the inter-stripe region (see detailed pictures and description in supplementary Fig. S5). In zebrafish, it was possible to show progressive waves via careful timing of fixation times (Holley et al. 2000). However, our attempts to do this also in Tribolium failed, because one can not achieve sufficient synchrony of the egg lays.
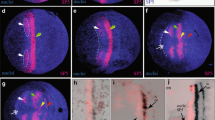
We have, therefore, assessed this question with other approaches. First, we collected a large number of pictures from the same stage of development and compared differences of expression in the growth zone. Figure 3a–c shows a series during the formation of hairy stripe #5. In this case, we have aligned the embryos with respect to the anterior borders of stripes #2 and #3, which can be considered to be in the zone of differentiated cells, i.e., should be static. Stripe #4 appears to retain a fixed anterior border in this case as well, while the posterior border shifts anteriorly, leaving a free space at the most posterior end (Fig. 3b). Stripe #5 develops in this free space, starting at the most posterior end (Fig. 3c). Given that the embryos appear to have grown during this time, one could interpret this also as a successive generation of stripes. The recession of the posterior border of stripe #4 could be interpreted as inter-stripe clearance, but could also be the result of cell intercalation (see “Discussion”).
Dynamics of hairy stripe formation in the growth zone. a–c In situ hybridization with the Tribolium hairy probe, d–i double staining with an engrailed antibody (brown). a to c shows three apparently successive stages during hairy stripe #5 formation. The embryos are aligned with respect to the anterior borders of stripes #2 and #3 (grey lines). d–i Successive stages during the formation of engrailed stripe 7, with an enlargement of the relevant section of the growth zone. Ordering of the stages is based on the appearance of the engrailed staining and the appearance of the enlargement of the growth zone
Double stainings with the segmental marker engrailed provide a similar picture. engrailed is expressed during formation of the segmental borders and is not expected to be dynamic itself. Figure 3e–i presents the series of stages from the beginning of the formation of the 6th to the 7th engrailed stripe, i.e. the equivalent of the formation of one segment. The distance between the last engrailed stripe and the neighboring hairy stripe does not seem to change during this time. Only the embryo itself elongates and the space for the emergence of the next engrailed stripe is thus formed.
A further possibility to investigate the question of moving expression waves is to express a long-lived gene product under the promotor of the gene in question. In zebrafish we could show that expression of gfp under the her1 promotor leads to a broad field of expression in the growth zone, since all cells express the gene at some point, although the shorter-lived RNA showed the expected stripes (Gajewski et al. 2003). Here we have used a gene construct that includes the regulatory region necessary for the expression of all eight Tc′h stripes fused to LacZ and a SV40 3′-UTR (Eckert et al. 2004) to study this question. Comparison of the expression of the construct at both mRNA and protein level with the endogeneous expression pattern provides no evidence for an expression wave in the growth zone (Fig. 4). Although the RNA and the protein expressed from the construct appear to be more stable than the endogeneous Tc-h RNA (compare stripes #2 and #3 in Fig. 4b,g,l), the resolution of stripes in the growth zone is highly comparable between the construct and the wild type situation.
a–e Expression pattern of the Tc-h-lacZ reporter gene construct KN8.8 (Eckert et al. 2004), as determined by in situ hybridization with the lacZ probe. The construct drives the expression of eight stripes, faithfully mimicking the wild type expression pattern. f–j Show an antibody staining for LacZ in the respective reporter line. Note that both the lacZ in situ (a–e) as well as the antibody staining (f–j) show formation of precise stripes (black arrows) with no staining in the inter-stripe region (white arrows), i.e., no fusions are detected as expected in the case of an expression wave k–o. Wild type Tc-h staining in comparable stages. Comparison of the expression in stripe #2 and #3 in b, g, and l shows higher stability of the lacZ transcripts and protein compared to the endogeneous hairy transcripts
Functional analysis of the gene Tc-hairy
Using RNAi analysis, Choe et al. (2006) found in their study on pair-rule genes in Tribolium that Tc-h does not appear to have a primary function in segmentation of the trunk region. Our results based on parental RNAi (pRNAi—Bucher et al. 2004) confirm this finding in principle, but we find apparently pair-rule-specific functions of Tc-h in the formation of the head segments.
pRNAi is usually very efficient in removing the gene product in the developing embryos, although one obtains phenotypic series of weaker effects depending on the age of the mothers (i.e., time after injection of the pupae). Injection of dsRNA as high as 2 μg/μl yielded the strongest phenotypes. Cuticles of the progeny of such treated females exhibit segmentation defects in the anterior part of the larval body, ranging from loss of the mandibular and labial segments in weak phenotypes up to loss of all segments anterior to the third thoracic segment (Fig. 5) in the strongest phenotype. However, analysis of the germband of these embryos reveals that these strongest phenotypes are partly due to a detachment of the remaining head structures (see below), i.e., they are not evidence for a role of Tc-h in the early specification of the most anterior head segments. Intermediate phenotypes display strong defects in gnathal segments, including either complete or partial loss of gnathal appendages like mandible, maxilla, and labium. Furthermore, left–right asymmetry of this phenotype was observed frequently, where appendages were lost, malformed, or strongly reduced on one side of the respective segment, but not the other. Analysis of the phenotypes obtained by injection of lower and higher dsRNA concentrations did correlate well with these phenotypes and no qualitative differences could be observed.
Cuticle preparations of Tc-h pRNAi knockdown embryos. a Wild type cuticle for comparison, lateral view. b–e Phenotypic series of cuticles, lateral view. In b, the distance between the antennae and the maxillae is strongly reduced, suggesting that the mandibular segment is missing (see also magnification in i). c–e Show successively stronger phenotypes were the entire head region is missing (c) and thoracic segments are affected (d–e). f, g Show magnifications of the head region in wild type (f) and intermediate Tc-h phenotypes (g) in a ventral view. In g, no mandibular structures are visible and the labium is strongly reduced. h–k Magnifications of the head region of a wild type (h) and three Tc-h pRNAi cuticles (i–k) in a lateral view. i Shows a magnification of the phenotype in b, note the distance between the antennae and the maxilla which lie directly adjacent in contrast to the wild type situation (h). In j, the labrum and the mandibles are absent whereas in k the labrum is present, but an antenna is formed only on one side, the maxillae are both present. Frequencies of phenotypes obtained with 2 μg/μl Tc-h dsRNA (b) 45%; (i–k) 29%; (c) 7%; (d) 3%; (e) 2%; 16% wild type, n = 114
Given the intricate regulation of Tc-h in the growth zone, as well as the conservation of this pattern in T. confusum, it seems surprising that Tc-h does not appear to have a function in the formation of the abdominal segments. Potentially, this could be due to a redundancy with the function of the homologue of deadpan, which is expressed in similar stripes as Tc-h (see above). However, pRNAi experiments with this gene did not yield abdominal segmentation phenotypes, neither in single injections, nor in double injections with Tc-h (data not shown). Hence, the lack of a trunk segmentation phenotype for Tc-h does not appear to be due to a redundancy with another hairy-like gene.
A pair-rule function of Tc-h at blastoderm stage
To better assess the function of Tc-h in anterior segmentation, we have used the segment polarity gene gooseberry (gsb) as a segmental marker. The first stripes of Tc-gsb are earlier expressed than Tc-en, which makes it particularly useful as marker. We find that in early embryos the stripes corresponding to the mandibular and labial segments are most strongly affected. They are either reduced, malformed, or even absent (Fig. 6f,g). In stronger phenotypes, the maxillary stripe can also be lost, but this is only observed at later stages (Fig. 6h). Hence, mandible and labium appear to be most sensitive to Tc-h function.
Analysis of the Tc-h knockdown phenotype during development. a–e Tc-gsb expression in wild type. Tc-gsb is expressed in the posterior part of the specified segments and serves as segmental marker. The positions of the mandibular and labial segments are marked with white arrows in a and b. f–j Tc-gsb expression in embryos depleted for Tc-h by pRNAi. The embryos depicted in f and g show disruption of Tc-gsb expression in the mandibular and labial segments (white arrows, compare to a and b). In older stages, the stripe corresponding to the maxillary segment is also affected (h–j), suggesting a secondary loss of this stripe, since a loss at early stages was never observed. Furthermore, head development appears to be significantly disturbed (compare i–j with d and e). Note the asymmetries of the phenotype in the embryos in h and j which we observed frequently
Depletion of Tc-h leads to induction of apoptosis in anterior segments
Although the cuticles of strong phenotypes lacked all head structures, we could not observe a loss of cephalic segments in the Tc-gsb stainings before germband retraction. However, head development appeared to be retarded and the tissue connection to the head was often only thin. We, therefore, performed a staining for apoptosis in order to determine whether tissue is lost during this phase. Staining with an antibody directed against the activated form of caspase3 allows the detection of apoptosis induction at early stages. There are indeed apoptosis-positive cells in the anterior region of extended germband stages (Fig. 7f–g). Interestingly, no apoptosis was detected along the ventral midline, although this is clearly seen in wild type embryos (Fig. 7c,d). We have, therefore, more carefully looked at the effects of Tc-h depletion on midline formation and find that there is indeed a visible phenotypic effect. In Tc-h knockdown embryos, the ventral midline appears broader and the cells lining the midline show abnormal morphology. They appear significantly larger compared to the wild type and extrude into the lateral regions of the embryo (Fig. 7i). Possibly, these are cells that would have undergone apoptosis in normal development. A comparable function is not known for Drosophila hairy, but interestingly, a role for the specification of vertebrate midline structures, derived from the Spemann–Mangold organizer and its respective counterpart, the dorsal shield, was found for different hairy homologues in zebrafish and Xenopus (Latimer et al. 2005; Murato et al. 2006).
Effects of Tc-h knockdown on apoptosis during development. Antibody staining against cleaved caspase3 (as apoptosis marker) in wild type (a and b, e and h) and in Tc-h pRNAi (c and d, f, g, and i). In wild type embryos apoptosis is only detected in a few cells in young stages (a). In older stages, apoptotic cells are found in the headlobes, the ventral midline and a few dispersed cells (b). Tc-h depleted embryos show patches of staining in the gnathal region as well as the first thoracic segment (c–d, f–g). Furthermore, disruption of the segments fusing the headlobes to the trunk region is seen, probably resulting in the detachment of all anterior segments during further development (f–g, black arrows). Note that no stained cells are detected along the ventral midline (h and i). Magnifications of the posterior segments of the embryos depicted in c and d. The midline appears wider and deeper in Tc-h knockdown embryos (black arrow) and large cells are found lining the border to the lateral regions of the embryo (white arrow, compare h and i). Segment identities abbreviated as follows: mandible, (md); maxilla. (mx); labium, (lb); T1–T3, thoracic segments
Regulation of target genes
The pair-rule gene ftz is negatively regulated by h in Drosophila (Ish-Horowicz and Pinchin 1987), an interaction that is likely to be direct (Dearolf et al. 1990). Although Tc-ftz is also expressed in a double-segmental pattern, previous analyses showed no evidence for an involvement of this gene in the segmentation process of Tribolium (Stuart et al. 1991). In Tc-h pRNAi embryos, the Tc-ftz stripes are initially formed, but do not resolve properly with ongoing development (Fig. 8f–j). In addition, the stripes are more persistent than in wild type embryos (Fig. 8i,j).
Effects of Tc-h knockdown on Tc-ftz expression. a–e Wild type expression of Tc-ftz and (f–j) in Tc-h pRNAi knockdowns. Tc-ftz is expressed in a pair-rule-like fashion complementary to Tc-h. Expression is detected in the maxillary segment of young germbands (a); thereafter, seven additional stripes appear sequentially, de novo near the tip of the expanding germband (b–e). In later stages, a second expression appears in the developing nervous system (e). In Tc-h knockouts, the anterior border of the first stripe seems unaffected (f) while the remaining stripes are formed, but appear to fuse with ongoing development (g–j). Interestingly, expression in the anterior region does not cease, as seen for the wild type expression. Instead, the expression persists throughout the segmentation process
Like Tc-ftz, Tc-prd is expressed in a pattern complementary to Tc-h, but in contrast to Tc-ftz, Tc-prd is required for Tribolium segmentation (Choe et al. 2006). In Tc-h knockdown embryos, the first Tc-prd stripe is initially formed as in wild type (Fig. 9g), while the second stripe appears directly adjacent to the first, consistent with a loss of the labial segment at blastoderm stage (Fig. 9h). The further stripes appear not to be affected by the Tc-h knockdown (Fig. 9i–k). In early germbands, a premature loss of the first stripe can be detected in part of the embryos (Fig. 9j), comparable to the loss of Tc-gsb expression observed in this segment. At later stages, no mandibular and labial appendages are seen in some of the embryos and the expression in the maxillae is reduced to one spot instead of the two seen in wild type embryos (Fig. 9m,n). Taken together, the results indicate a specific loss of the mandibular and labial segment, which fits with the observations at the cuticle level. The missing spot of Tc-prd expression in the maxilla may be a result of the secondary function of Tc-h.
Effects of Tc-h knockdown on Tc-prd expression. a–f, m Wild type expression of Tc-prd and (g–l, n) in Tc-h pRNAi knockdowns. Expression of Tc-prd starts as a circumferential stripe in the blastoderm in the maxillary segment, which is followed by a second stripe of expression in the segment corresponding to T1 before the formation of the germ rudiment (a–b). The following stripes appear de novo at the anterior border of the growth zone (c–f). Shortly after the appearance of the stripes, they split into a segmental expression pattern. At the end of segmentation Tc-prd is strongly expressed in the mandible and the maxilla (m). In Tc-h knockdown embryos, the first Tc-prd stripe appears normally in the blastoderm (g), whereas the second stripe is formed directly adjacent to the first (h), suggesting a loss of the intermediate labial segment. During further development, these stripes do not split as seen in wild type. Instead, a weaker stripe is seen in the region of the presumptive labial segment (compare i and c, white arrow). In some cases, a loss of expression in the maxillary segment is observed during further germband growth (j, black arrow). The remaining stripes appear unaffected (k–l). m–n Show magnifications of the head region in wild type (m) and Tc-h depleted embryo (n). Expression in the mandible is not detected and no mandibular nor labial structures are formed (compare n and m)
Discussion
The function of hairy in Tribolium remains enigmatic, although our results clarify several points. The apparent lack of function during the trunk segmentation process (Choe et al. 2006) is not due to a redundancy with another hairy homologue in Tribolium. The expression of Tc-h during trunk segmentation could alternatively have been due to an accidental enhancer capture from another pair-rule gene. However, this can now also be ruled out. First, there is no other pair-rule gene homologue close to Tc-h in the genome sequence (the neighboring genes are the “signal recognition particle receptor beta subunit” and the “nicotinic acetylcholine receptor subunit alpha2”) and second, the expression is conserved in T. confusum, which would be very unlikely, if it would have no function.
Interestingly, expression and function of Tc-h during blastoderm is compatible with a pair-rule function. The mandibular and the labial segments correspond to the anterior parts of the first and second Tc-h stripe at blastoderm and they are the ones that are most sensitive to a loss of function of Tc-h. This is in line with a classic pair-rule function and is also not in contrast to the results by Choe et al. (2006) since the formation of the head segments was not specifically addressed in this study. Our analysis of the hunchback function in Tribolium (Marques-Souza et al. 2008) has also suggested that the patterning of the head segments should be seen separately from the trunk segments. This is also a classical conclusion from comparative morphology (reviewed in Tautz 2004). Hence, it seems possible that only the head segment specification is a conserved feature of hairy function between Drosophila and Tribolium. This would imply that the striped expression of hairy, although conserved in other Arthropoda, might have been recruited to the trunk segmentation process only in the lineage leading to Drosophila. We can only speculate about the specific role of the hairy stripe formation in the trunk. Most likely, it is related to some differentiation process in the developing nervous system.
The strong phenotypes in Tc-h knockdowns lead to a loss of adjacent segments in the head and thorax region, i.e., this goes beyond a pair-rule phenotype. This phenotype appears to involve specific cell death in the respective region, although the region that is affected in the strongest cuticle phenotypes is larger than the region where we observe cell death (i.e., it includes part of the thorax). This raises the possibility that Tc-h may have an organizer function in this region, which would determine the fate of surrounding cell groups. It should be noted that the first Tc-h stripe remains expressed much longer than the subsequent stripes, a feature that is also conserved in T. confusum. Interestingly, this expression overlaps with a specific expression of Tc-delta in this region (supplementary Fig. S4) supporting the notion of signaling processes being activated there. In any case, it appears that this possible organizer function occurs subsequently to the segmentation function.
The emergence of the Tc-h stripes in the growth zone is superficially similar to the oscillatory expression of hairy homologues in vertebrates, but none of our experiments supports the notion that there is an expression wave across the growth zone. The fact that the more long-lived lacZ reporter gene shows essentially the same stripe pattern as the wild type expression is probably most telling. If the expression would be a moving wave, the interstripe cells should also express lacZ, i.e., the resolution of the stripes should be blurred. This is not observed, although the expression appears to be somewhat dynamic with respect to the apparent clearance of stripes in the growth zone (Fig. 3). Given that no specific cell-division activity is apparent in this region (supplementary Fig. S6), we have to conclude that cell migration and intercalation are likely to play a role in the generation of this pattern. Similar as in vertebrates, the Tribolium embryo undergoes a convergent extension process during germband growth (compare supplementary Fig. S5). The cellular interactions and movement patterns during convergent extension processes are still not fully understood (see Keller et al. 2008 for a recent discussion), but it appears unlikely that they are involved in the generation of the stripes in the growth zone. Hence, further work will be required to understand this patterning mechanism and the general process of segment formation in short-germband embryos.
References
Aranda M (2006) Functional analysis of a homologue of the pair-rule gene hairy in the short-germ beetle Tribolium castaneum. PhD thesis, University of Cologne
Baumgartner S, Noll M (1990) Network of interactions among pair-rule genes regulating paired expression during primordial segmentation of Drosophila. Mech Dev 33:1–18
Berghammer A, Bucher G, Maderspacher F, Klingler M (1999) A system to efficiently maintain embryonic lethal mutations in the flour beetle Tribolium castaneum. Dev Genes Evol 209:382–389
Bier E, Vaessin H, Younger-Shepherd S, Jan LY, Jan YN (1992) deadpan, an essential pan-neural gene in Drosophila, encodes a helix–loop–helix protein similar to the hairy gene product. Genes Dev 6:2137–2151
Bucher G, Scholten J, Klingler M (2004) Parental RNAi in Tribolium (Coleoptera). Curr Biol 12:85–86
Carroll SB, Laughon A, Thalley (1988) Expression, function, and regulation of the hairy segmentation protein in the Drosophila embryo. Genes Dev 2:883–890
Choe CP, Miller SC, Brown SJ (2006) A pair-rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. Proc Natl Acad Sci USA 103:6560–6564
Dearolf CR, Topol J, Parker CS (1990) Transcriptional regulation of the Drosophila segmentation gene fushi tarazu (ftz). BioEssays 12:109–113
Eckert C, Aranda M, Wolff C, Tautz D (2004) Separable stripe enhancer elements for the pair-rule gene hairy in the beetle Tribolium. EMBO Rep 5:638–642
Gajewski M, Sieger D, Alt B, Leve C, Hans S, Wolff C, Rohr KB, Tautz D (2003) Anterior and posterior waves of cyclic her1 gene expression are differentially regulated in the presomitic mesoderm of zebrafish. Development 130:4269–4278
Gajewski M, Elmasri H, Girschick M, Sieger D, Winkler C (2006) Comparative analysis of her genes during fish somitogenesis suggests a mouse/chick-like mode of oscillation in medaka. Dev Genes Evol 216:315–332
Giudicelli F, Lewis J (2004) The vertebrate segmentation clock. Curr Opin Genet Dev 14:407–414
Gutjahr T, Frei E, Noll M (1993) Complex regulation of early paired expression: initial activation by gap genes and pattern modulation by pair-rule genes. Development 117:609–623
Holley SA, Geisler R, Nüsslein-Volhard C (2000) Control of her1 expression during zebrafish somitogenesis by a Delta-dependent oscillator and an independent wave-front activity. Genes Dev 14:1678–1690
Ingham PW (1988) The molecular genetics of embryonic pattern formation in Drosophila. Nature 335:25–34
Ish-Horowicz D, Pinchin SM (1987) Pattern abnormalities induced by ectopicexpression of the Drosophila gene hairy are associated with repression of ftz transcription. Cell 51:405–415
Jürgens G, Wieschaus E, Nüsslein-Vollhard C (1984) Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. Roux Arch Dev Biol 193:283–295
Keller R, Shook D, Skoglund P (2008) The forces that shape embryos: physical aspects of convergent extension by cell intercalation. Phys Biol 5:15007
Klingler M, Gergen JP (1993) Regulation of runt transcription by Drosophila segmentation genes. Mech Dev 43:3–19
Latimer AJ, Shin J, Apple B (2005) her9 promotes floor plate development in zebrafish. Dev Dyn 232(4):1098–1104
Macdonald PM, Struhl G (1986) A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature 324:537–545
Marques-Souza H, Aranda M, Tautz D (2008) Delimiting the conserved features of hunchback function for the trunk organization of insects. Development 135:881–888
Murato Y, Yamaguti M, Katamura M, Cho KW, Hashimoto C (2006) Two modes of action by which Xenopus hairy2b establishes tissue demarcation in the Spemann–Mangold organizer. Int J Dev Biol 50:463–471
Nüsslein-Volhard C, Wieschaus E (1980) Mutations affecting segment number and polarity in Drosophila. Nature 287:795–801
Rida PCG, Le Minh N, Jiang JY (2004) A Notch feeling of somite segmentation and beyond. Dev Biol 265:2–22
Rushlow CA, Hogan A, Pinchin SM, Howe KM, Lardelli M, Ish-Horowicz D (1989) The Drosophila hairy protein acts in both segmentation and bristle patterning and shows homologue to N-myc. EMBO J 8:3095–3103
Sommer RJ, Tautz D (1993) Involvement of an orthologue of the Drosophila pairrule gene hairy in segment formation of the short germ-band embryo of Tribolium (Coleoptera). Nature 361:448–450
Stollewerk A, Schoppmeier M, Damen WGM (2003) Involvement of Notch and Delta genes in spider segmentation. Nature 423:863–865
Stuart JJ, Brown SJ, Beeman RW, Denell RE (1991) A deficiency of the homeotic complex of the beetle Tribolium. Nature 350:72–74
Tautz D (2004) Segmentation. Dev Cell 7:301–312
Tautz D, Pfeifle C (1989) A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98:81–85
Tribolium Genome Sequencing Consortium (2008) The genome of the model beetle and pest Tribolium castaneum. Nature 452:949–955 doi:10.1038/nature06784
Tsai C, Gergen P (1995) Pair-rule expression of the Drosophila fushi tarazu gene: a nuclear receptor response element mediates the opposing regulatory effects of runt and hairy. Development 121:453–462
Acknowledgements
This work was supported by the DFG (SFB 572 and SFB 680). H. M. was a fellow of the International Graduate School of Genetics and Functional Genomics in Cologne.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Roth
Manuel Aranda and Henrique Marques-Souza contributed equally to this work.
Electronic supplementary material
Below is the linked to the electronic supplementary material
supplementary Fig. S1
Expression of Tc’deapan in Tribolium (DOC 1170 kb)
supplementary Fig. S2
Sketch of conserved blocks between the T. castaneum and T. confusum genomic regions around hairy. In addition to the conserved exons (in red), there are several upstream regions with more than 50% identity (green blocks). Note the inversion of the most upstream block (DOC 40 kb)
supplementary Fig. S3
Expression of hairy in T. confusum during development (DOC 3529 kb)
supplementary Fig. S4
Expression of genes from the Delta/Notch signaling pathway in Tribolium. Notch is broadly expressed in more or less all cells during early stages. Delta expression is restricted to an anterior stripe, the midline with a stronger expression in the growth zone at early stages, and the nervous system at late stages. Serrate and Fringe are expressed only at late stages. None of these patterns suggest a possible involvement of Delta–Notch signaling in hairy stripe formation in the growth zone (DOC 730 kb)
supplementary Fig. S5
Expression of Tc′h in wild type. Blastoderm satges are aligned with the anterior to the left and the ventral side up, except for (b) which shows a ventral view. Germband stages are aligned with anterior to the left and numbers indicate the respective stripes. a–d Expression during blastoderm stage. Expression of Tc’h starts in a broad, circumferential stripe of expression positioned in the centre of the early blastoderm (a). b Expression of the second stripe starts as a posterior cap which slowly condenses at the anterior border and narrows down to become stripe #2 while expression retracts from the posterior pole at the same time (c–d). In e, gastrulation begins and all three blastodermal stripes are expressed. (f–l) Shows the formation of the stripes #3–#5 in detail. The germband in (f) shows the expression of stripe #1–#3, note that expression is not detected at the most posterior end of the embryo. In G, the posterior border of stripe #3 is completely refined while expression in the growth zone covers the entire posterior part of the embryo. In h–j, expression retracts in the most posterior part of the growth zone while condensing at the anterior border at the same time, forming stripe #4. Note that the expression at the posterior border seems to be highly dynamic while the anterior border is always clearly refined. In k, the cycle starts all over leading to the formation of stripe #5. l The remaining three stripes are formed in an identical fashion. m–r In older stages, expression is detectable in the presumptive nervous system in form of spots in the lateral regions of the embryo and in the headlobes (DOC 2514 kb)
supplementary Fig. S6
DAPI staining of the growth zone to trace cell-division patterns. The pictures are taken from the series of double stainings with hairy and engrailed (compare Fig. 3). Metaphase stages are rare during germband growth and also not concentrated at the most posterior end. Hence, cell movement is more likely to cause the growth process (DOC 2016 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Aranda, M., Marques-Souza, H., Bayer, T. et al. The role of the segmentation gene hairy in Tribolium . Dev Genes Evol 218, 465–477 (2008). https://doi.org/10.1007/s00427-008-0240-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-008-0240-1