Abstract
Mutations in the gene OCA2 are responsible for oculocutaneous albinism type 2, but polymorphisms in and around OCA2 have also been associated with normal pigment variation. In Europeans, three haplotypes in the region have been shown to be associated with eye pigmentation and a missense SNP (rs1800407) has been associated with green/hazel eyes (Branicki et al. in Ann Hum Genet 73:160–170, 2009). In addition, a missense mutation (rs1800414) is a candidate for light skin pigmentation in East Asia (Yuasa et al. in Biochem Genet 45:535–542, 2007; Anno et al. in Int J Biol Sci 4, 2008). We have genotyped 3,432 individuals from 72 populations for 21 SNPs in the OCA2-HERC2 region including those previously associated with eye or skin pigmentation. We report that the blue-eye associated alleles at all three haplotypes were found at high frequencies in Europe; however, one is restricted to Europe and surrounding regions, while the other two are found at moderate to high frequencies throughout the world. We also observed that the derived allele of rs1800414 is essentially limited to East Asia where it is found at high frequencies. Long-range haplotype tests provide evidence of selection for the blue-eye allele at the three haplotyped systems but not for the green/hazel eye SNP allele. We also saw evidence of selection at the derived allele of rs1800414 in East Asia. Our data suggest that the haplotype restricted to Europe is the strongest marker for blue eyes globally and add further inferential evidence that the derived allele of rs1800414 is an East Asian skin pigmentation allele.
Similar content being viewed by others
Background
Many genes have been associated with normal variation in human pigmentation (Sturm 2009; Sturm and Larsson 2009). Of those, OCA2 [MIM 611409], named for an abnormal pigmentation phenotype, oculocutaneous albinism type II (OCA2 [MIM 203200]), is a large gene extending over 300 kb on chromosome 15. OCA2 encodes the protein P, a transmembrane protein, and has been shown to play a role in pigmentation in both humans and mice (Frudakis et al. 2003). In humans, it has been implicated in iris, skin, and hair pigmentation (Duffy et al. 2007; Sturm et al. 2008; Kayser et al. 2008; Sulem et al. 2007). The exact function of P is unknown though it has been suggested to process and traffic tyrosinase, regulate melanosomal pH, or regulate glutathione metabolism (Toyofuku et al. 2002; Staleva et al. (2002); Sturm et al. 2001; Edwards et al. 2010).
Mutations in OCA2 are known to cause oculocutaneous albinism type 2. However, the gene is also known to play a role in variation in normal pigmentation. In European populations, it is primarily associated with blue irises. Several sites in and around OCA2 have been reported to be the functional variant or to be tightly linked to the functional variant leading to blue eyes. These sites include a three-SNP haplotype (rs4778138, rs4778241, rs7495174) and four individual SNPs, rs1129038, rs12913832, rs916977, and rs1667394 (Duffy et al. 2007; Sturm et al. 2008; Kayser et al. 2008; Sulem et al. 2007; Mengel-From et al. 2010; Walsh et al. 2010). Four of the SNPs (rs1129038, rs12913832, rs916977, rs1667394) are actually located in introns of the Hect Domain and RCC1-like Domain 2 (HERC2 [MIM 605837]), which are located 10 Kb upstream of OCA2. These are thought either to be located in or near an upstream regulatory region of OCA2 or to be in linkage disequilibrium (LD) with functional elements in HERC2 and affect a possible HERC2 regulation of OCA2. The actual function of HERC2 is unknown but it shows homology to known E3 ubiquitin-protein ligases. One of the HERC2 SNPs (rs1667394) has been associated with blond hair in Europeans (Sulem et al. 2007). Specific polymorphisms and the haplotypes are illustrated in Fig. 1; all 21 SNPs studied are listed in Table 2. The derived allele of another SNP at OCA2, rs1800407, has been associated with green/hazel eyes in Europeans (Branicki et al. 2009). Rs1800407 is an arginine to glutamine missense mutation (Arg419Gln) found in exon 13 of the OCA2 gene. Sturm et al. (2008) concluded that the derived allele of rs1800407 increased the penetrance of the blue eye phenotype associated with the derived allele of rs12913832.
The derived allele at a missense SNP (rs1800414, His615Arg) in exon 19 of OCA2 has been reported to be specific to East Asia (Yuasa et al. 2007; Anno et al. 2008). Edwards et al. (2010) showed an association between the derived allele of rs1800414 (C, 615Arg) and lighter skin pigmentation in a sample of individuals of East Asian ancestry from Canada and confirmed their results using an independent sample of Han Chinese.
Here we present our results on the global distributions of haplotypes and specific SNPs in the region of OCA2 and HERC2, genes that have been implicated in pigmentation variation in Europeans and East Asians. We also examine the LD between the SNPs and haplotypes of interest. Finally, we use long-range haplotype tests to show that OCA2 is or has been under selection in Europe and the derived allele of rs1800414 is, or has been, under selection in East Asia.
Materials and methods
Populations
We have typed 3,432 individuals from a global sample of 73 populations. The populations represent regions of Africa (13 populations), Southwest Asia (5), Europe (16), Siberia (3), South Central Asia (6), East Asia (17), the Pacific Islands (4), North America (4), and South America (5) (Table 1). Where available we also included data from the Human Genome Diversity Panel (Li et al. 2008b; Jakobsson et al. 2008). We combined certain smaller closely related HGDP population samples to form larger samples for our analyses (see Table 1).
DNA was extracted from lymphoblastoid cell lines for 57 of the population samples. The cell lines were established and/or maintained using common techniques described elsewhere (Anderson and Gusella 1984) in the lab of Kenneth K. and Judith R. Kidd at Yale University. Some cell lines were established by the Coriell Cell Repositories and by the National Laboratory for the Genetics of Israeli Populations at Tel Aviv University. The DNA for the 15 other population samples was obtained as DNA only from colleagues or the Coriell Cell Repositories (see Supplemental data). All samples were collected with informed consent by participants and with approval by all relevant institutional review boards.
Whole genome amplification
For the 15 DNA-only population samples, the DNAs were initially whole genome amplified using multiple displacement amplification (MDA), as described in Li et al. (2008a).
SNP typing
We typed all of the implicated SNPs as well as others for a total of 21 SNPs spanning a total of 398,549 bp (Table 2) in our 72 population samples. Nine of the SNPs (rs4778138, rs4778241, rs7495174, rs1129038, rs12913832, rs916977, rs1667394, rs1800407, rs1800414) were chosen because of their previous association with pigmentation; the remainder was chosen based on allele frequencies in European populations from the Applied Biosystems SNP catalogue and to bring up the average coverage to one SNP for every 20 kb. SNPs were typed using Applied Biosystems TaqMan® assays performed in 384-well plates using ~50–100 ng of DNA per well. We analyzed the SNP typing results using the ABI Prism Sequence Detection System.
Analyses
In addition to the data we generated, where available, we included data from the HapMap and the HGDP 650 k panel for rs4778138, rs4778241, rs7495174, rs12913832, and rs1667394 (Li et al. 2008b; Jakobsson et al. 2008). We omitted the HGDP data for those individuals who are part of our laboratory’s cell line collection and typed in our laboratory because we have larger sample sizes. All haplotypes were estimated using fastPHASE, and frequency maps were created using Surfer (ver 7) (Scheet and Stephens 2006). LD was calculated and LD figures were generated using HAPLOT with default parameters (Gu et al. 2005). For the selection studies we used relative extended haplotype homozygosity (REHH) and where applicable normalized haplosimilarity (nHS) (Sabeti et al. 2006; Hanchard et al. 2006). REHH and nHS are both based on the logical assumption that a variant under selection will rise to high frequency quickly before recombination has time to break down the extended haplotype on which the variant initially arose. In contrast, a neutral variant will take longer to reach a high frequency, allowing the extended haplotype time to be degraded by recombination. For the REHH test, a core haplotype containing the variant of interest is selected, an extended haplotype homozygosity score is then determined for each of the remaining SNPs moving outward from the core haplotype in each direction. Relative EHH scores weighted for allele frequency are then calculated for each of the non-core SNPs for each allele of the core haplotype, the scores of the SNP(s) furthest from the core are then tested for significance using 1,000 neutral simulations. nHS uses a moving window to determine a z-score for the least frequent allele of all SNPs in the dataset; again each z-score is compared to 1,000 datasets simulated under neutral conditions to determine if any show evidence of selection. Since nHS can only calculate a z-score for the least frequent allele of a given variant, it was only used when the allele of interest had a frequency <0.5. REHH and nHS was calculated using pselect (Han et al. 2007). Simulated data were created using Hudson’s ms (Hudson 2002). Two demographic models were used; the first was a model of a constant population size, and the second was a model of a bottleneck followed by an exponential expansion (a population starting 4,000 generations ago with a bottleneck occurring 1,600 generations ago and dropping the effective population size from 10,000 to 2,000 followed by an exponential expansion starting 400 generations ago leading to a population size of 100,000).
Results
SNPs
The allele frequencies for all 21 SNPs in all 73 population samples we genotyped are available in ALFRED (http://alfred.med.yale.edu) under the OCA2 and HERC2 loci or directly for each SNP by using the rs number in Table 2 as a keyword. As shown in Table 2, almost all of the SNPs had very large global allele frequency ranges, though for most SNPs the highest derived allele frequencies are found in Europeans. Other than rs1800407, with a range from 0.890 to 1.000 for the ancestral allele, the global allele frequency ranges are all above 0.7.
Blue-eye associated haplotypes
The three haplotype systems we define here are shown in Fig. 1 and Table 3. Duffy et al. (2007) previously identified a three-SNP haplotype system (rs4778138, rs4778241, and rs7495174) associated with blue eyes; for the purpose of this paper, we will refer to this system as BEH1, blue-eye associated haplotype #1. The blue-eye associated allele of BEH1 is ACA, the fully derived haplotype. Sturm et al. (2008) reported that rs12913832 is associated with blue eyes. Since rs1129038 is in nearly complete LD with rs12913832 in all populations, we defined these two SNPs as a haplotype system referred to as BEH2, blue-eye associated haplotype #2. The blue-eye associated allele of BEH2 is TG, both derived alleles. In the HGDP populations, BEH2 will consist of rs12913832 only since rs1129038 is not present in that dataset. We also typed an SNP that occurs between rs12913832 and rs1129038; however, it has not been associated with pigmentation, and is monomorphic on the blue-eye associated allele of BEH2 and was therefore not included in BEH2. Two other SNPs, rs916977 and rs1667394, have previously been associated with blue eyes (Kayser et al. 2008; Sulem et al. 2007). In our data, with the exception of a low frequency haplotype in Africa, rs916977 and rs1667394 are in nearly complete LD. Therefore, we treat them as another haplotype system, BEH3, blue-eye associated haplotype #3. The blue-eye associated allele of BEH3 is CA, again the derived haplotype. In the HGDP populations BEH3 will consist of rs1667394 only since rs916977 is not present in the data set.
Geographic distributions of haplotypes
The distributions of the blue-eye associated alleles at the three haplotyped systems are presented in Fig. 2, each haplotype in contour plots, and all three grouped by population in a histogram. The actual frequencies are presented in supplemental material and in ALFRED. The alleles associated with blue eyes at all three BEH blue-eye associated haplotypes have their highest frequencies in Northwestern Europe, and the TG allele at BEH2 is essentially observed only in Europe; the ACA allele of BEH1 and the CA allele at BEH3 are at their highest frequencies in Europe, particularly in Northern and Western Europe, and have much lower frequencies elsewhere. In most of Central and East Asia, these alleles have frequencies of <20% but reach frequencies of 40% and higher in the Americas.
Global frequencies of blue-eye associated haplotypes. This figure shows the distributions of the blue-eye associated allele/haplotype at the respective BEH1 (a), BEH2 (b), and BEH3 (c) genetic systems graphed on a world map, as well as a comparison of the frequencies in a bar graph (d). In part d, the associated alleles are represented in yellow at BEH1, in blue at BEH2, and in red at BEH3. Here we see that the blue-eye associated allele of BEH2 is mostly limited to Europe, whereas the blue-eye associated alleles of BEH1 and BEH3 are found globally. The populations are divided by regional group on the x-axis as follows: Africa (yellow), Southwest Asia (green), Europe (blue), Central Asia (orange), Pacific Islands (purple), East Asia (red), and Native Americans (teal)
Geographic distribution of the derived allele of rs1800407
The derived allele of rs1800407 is relatively rare compared to the blue-eye associated alleles of the three BEHs. The derived allele frequencies of rs1800407 are presented in Fig. 3. The derived allele is mostly restricted to Europe (0–11%), Southwest Asia (0–9.4%), and Central Asia (0–9.3%). Outside of this region, the derived allele is found in African Americans (1.7%), San Francisco Chinese (0.9%), the Arizona Pima (1.0%), and the Maya (3.9%).
Global distribution of the derived allele (T) of rs1800407. This figure shows the derived-allele frequencies of rs1800407. The derived allele is primarily restricted to Europe, Southwest Asia, and Central Asia, and has a maximum allele frequency of 11% in any given population sample. The populations are divided by regional group on the x-axis as follows Africa (yellow), Southwest Asia (green), Europe (blue), Central Asia (orange), Pacific Islands (purple), East Asia (red), and Native Americans (teal)
The T allele of rs1800407 has also been associated with blue-eye penetrance (Sturm et al. 2008). We estimated haplotype frequencies for haplotypes containing rs1800407 and the three BEHs (supplemental Fig. 1). The first observation is that the blue-eye associated alleles of the three BEHs are much more common than the derived allele of rs1800407. At BEH1, the T allele of rs1800407 most commonly occurs with the AAA allele and not the ACA allele that has been associated with blue eyes. The T allele with the ACA blue-eye associated allele is the second most common combination. Other combinations occur but they are rare. The T allele of rs1800407, when seen, is commonly paired with the blue-eye associated TG allele at BEH2 only in Northern and Eastern Europeans. This association may explain the increased blue-eye penetrance seen by Sturm et al. (2008) as a type of ascertainment effect. Elsewhere the T allele is more likely to be found paired with the CA allele. We see a similar pattern at BEH3 as we see at BEH2. The blue-eye associated CA allele of BEH3 commonly pairs with the T allele only in Northwestern and Eastern Europe and the TG allele is its most common partner elsewhere.
Geographic distribution of the derived allele of rs1800414
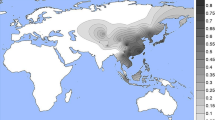
Our data confirm that the putative light skin allele of rs1800414 (C) is found almost exclusively in East and Southeast Asia, at frequencies ranging from 0 to 76% (Fig. 4) at higher levels in eastern East Asia (62–76.1%) compared with Southeast Asia (0–54.3%) and Western China (15.5–37.5%). Outside of East and Southeast Asia, the C allele is only found in low frequencies in the Adygei, Chuvash, and Hungarians in Europe (>1–3.6%), the Yakut in Siberia (8.8%), and the Micronesians in the Pacific Islands (4.2%).
Global rs1800414 derived-allele distribution and frequencies. This figure shows the distribution of the derived allele of rs1800414 interpolated on a world map (a) and as a bar graph (b). The derived allele is essentially restricted to East Asia, with the highest frequencies in Eastern East Asia, midrange frequencies in Southeast Asia, and the lowest frequencies in Western China and some Eastern European populations
Haplotypes and LD
We calculated pairwise r 2 for all 21 SNPs and illustrate regions of high LD using the HAPLOT program (Fig. 5). On average, globally we see two regions of high LD, though the sizes of each of these regions vary by population group. In Africa, the first region encompasses SNP 4 (rs12914687) through SNP 7 (rs2015343) and the second region encompasses SNP 16 (rs7494942) through SNP 21 (rs1667394). In Southwest Asia and Europe, both high LD regions are larger and the first is composed of SNP 3 (rs11074314) through SNP 8 (rs4778136), and the second is composed of SNP 12 (rs4778138) through SNP 21 (rs1667394). In Central Asia and the Pacific, the first region is the same as in Africa and the second region is the same as in Southwest Asia and Europe. In East Asia, the first high LD region extends from SNP 2 (rs1800414) to SNP 9 (rs746861) and the second region extends from SNP 10 (rs7170869) to SNP 21 (rs1667394). We actually see three regions of high LD in Native Americans, the first from SNP 3 (rs11074314) to SNP 8 (rs4778136), the second from SNP 9 (rs746861) to SNP 12 (rs4778138), and the third from SNP 18 (rs3935591) through SNP 21 (rs1667394). In Europe, the second region covers all three BEHs, and in East Asia, the first region includes rs1800414.
LD at OCA2 and HERC2. This figure shows the LD in the OCA2/HERC2 region in 55 populations. SNPs 1–21 are ordered as in Table 2. A region of high LD is represented by red arrows using the default parameters in the agglomerative algorithm in HAPLOT (Gu et al. 2005): A region of high LD starts at r 2 = 0.4 and is extended as long as the average r 2 ≥ 0.3. The minimum r 2 for inclusion in a block is 0.1. If LD cannot be calculated for a SNP (e.g., it is fixed in that particular population), then a white space in the arrow is shown. On average, there are two regions of high LD, one near the East Asian “light skin” SNP (rs1800414) and one in the BEH region. The smallest regions are in Africa whereas the largest regions are in East Asia. In the Americas, there are three regions, one near rs1800414, one at BEH1, and one at BEH3
Since the blue-eye associated alleles at all three BEHs are concordant in Europe and fall into that same high LD region in Europe, we analyzed the haplotypes of all seven SNPs together (Fig. 6). In this data set, we see that the TG allele BEH2 always occurs on chromosomes that have the CA allele of BEH3 and almost always occurs on chromosomes with the ACA allele of BEH1. The ACA allele of BEH1 and the CA allele of BEH3 also usually occur on the same chromosomes; however, outside of Northwestern and Eastern Europe they do not always occur on chromosomes with the TG allele of BEH2. Whenever one of the blue-eye associated alleles does occur on a chromosome by itself, it is most likely to be the CA allele of BEH3.
Haplotypes of the three BEHs. This figure shows the three BEHs as a single haplotyped system. The TG allele of BEH2 always occurs with the CA allele of BEH3 and usually occurs with the ACA allele of BEH1 (yellow). The CA BEH3 and ACA BEH1 alleles, however, do not always occur with the TG allele of BEH2. The ACA BEH1 allele and the CA BEH3 allele also usually occur together (pink and yellow)
We also looked at the haplotypes of the seven SNPs that compose the first high LD region in East Asians with respect to the derived allele of rs1800414 (Fig. 7). Here we see the derived allele of rs1800414 occurs on three haplotypes, though a vast majority occurs on a single haplotype (CACCACT). Of the remaining two haplotypes containing the derived allele of rs1800414, one differs from the most common haplotype at the last site and the other differs at the final four sites.
Haplotypes containing the derived allele of rs1800414 in East Asians. This figure shows a seven-SNP haplotype in the “light skin” region of OCA2 in East Asians. The seven SNPs were chosen based on the first region of high LD in East Asians from Fig. 4. The C allele of rs1800414 is seen on three haplotypes, one of which (blue) accounts for a large majority of the chromosomes. The next most common haplotype (red) differs from the most common (blue) only at the seventh site. The least common (yellow) differs from the blue at the final four sites
Selection
We tested all five pigmentation regions for evidence of positive selection using REHH. For the “light skin” allele at rs1800414 and the blue-eye penetrance allele at rs1800407 we tested the REHH value at rs1667394, for the blue-eye associated haplotypes at BEH1 we tested at SNPs rs2703969 and rs1667394, and at BEH2 and BEH3, we tested at rs2703969. These SNPs were chosen to test for significance because they were the most distant SNPs from their respective core and fell the ideal distance away according to the protocol described by Sabeti et al. 2006. Since REHH requires a core haplotype with multiple alleles for comparison, rs1800414 was included in a haplotype with rs11074314 and rs12914687. The C allele of rs1800414 only occurred on a single allele of this haplotype. We also added an extra SNP to BEH2 (rs7494942) and BEH3 (rs7170852) haplotypes. Again, the alleles of interest only occurred on one haplotype. We tested all the populations grouped by region: Africa, Southwest Asia, Europe, East Asia, and America. In the European sample using the constant population size simulation model, we see the strongest signal for selection at the TG allele of BEH2 (Fig. 8). At the ACA allele of BEH1 and the CA allele of BEH3, the REHH scores are weakly significant and just over the 95th percentile; however, both regions are within the false positive grouping of the simulated data. We also subdivided Europe into three groups: Southern Europe, Eastern Europe, and Northwestern Europe. In Southern Europe, the TG allele at BEH2 has a strongly significant REHH score, at BEH3 the CA allele is weakly significant, and there is no evidence of selection at BEH1. In Eastern Europe, the evidence for selection is again the strongest at the TG allele BEH2; there is no evidence of selection to the centromeric side of BEH1, and weak evidence for selection at the CA allele of BEH3 and to the telomeric side of the ACA allele of BEH1. In Northwestern Europe, the TG allele of BEH2 once again has the strongest signal for selection, the centromeric side of the ACA allele of BEH1 and the CA allele of BEH3 are very weakly significant, and the telomeric side of BEH1 shows no significant evidence of selection. In Southwest Asia, there are significant REHH values at the TG allele of BEH2 and the CA allele BEH3. As in Europe, the BEH2 signal of selection is strong, whereas the BEH3 signal is barely significant (see supplemental Fig. 2). We confirmed these results using a bottleneck followed by an exponential expansion model and saw the same results (supplemental Fig. 3). In fact, though the bottleneck with expansion model had a different distribution of allele frequencies (more high frequency alleles and fewer midrange frequency alleles compared to the constant population size model) the 95th percentile line remained the same. Since the frequencies of blue-eye associated alleles of the SNPs that compose BEH2 are <50% in Southern Europe and Southwest Asia, we were able to confirm these results using a second LRH test, normalized haplosimilarity (nHS). Again, we see strong evidence of selection at the two BEH2 SNPs in Southern Europe and Southwest Asia using the nHS test (Fig. 9). No evidence of selection was seen at rs1800407 (supplemental Fig. 4).
Relative extended haplotype homozygosity test at the blue-eye associated haplotypes in Europe. This figure shows graphs of the REHH (a, c, e, g) and the significance tests (b, d, f, h) for the three blue-eye associated haplotypes in Europe. a, b Graphs for the SNPs centromeric to BEH1. In the significance test graphs, the cyan points are the REHH results from 1,000 simulations under the constant population size neutral model. The ACA allele is right at the 95th percentile and well within the area of false positives. c, d Graphs for the SNPs telomeric to BEH1. Again, the ACA allele is right at the 95th percentile line and well within the area of false positives. e, f Graphs for the SNPs centromeric to BEH2. Here we see the TG allele is above the 95th percentile line suggesting a strong signal of selection at this locus. g, h Graphs for the SNPs centromeric to BEH3. The CA allele is also above the 95th percentile line but the signal is not as strong as for BEH2
nHS at OCA2/HERC2 in Southern Europeans. This figure shows the results for a normalized haplosimilarity test in Southern Europeans. Southern Europeans were chosen because they are the only group of Europeans in whom any of the frequencies of blue-eye associated alleles of the three blue-eye haplotypes falls below 0.50, a requirement for this test to detect selection. The cyan points represent the result of 1,000 simulated populations under the neutral constant population size model and the blue points represent the data at OCA2/HERC2. The only two points that show a significant result are the two SNPs that compose BEH2
In East Asia we see strong evidence for selection at the C allele of rs1800414 using the REHH test in both the constant population size model (Fig. 10a, b) and the bottleneck with an expansion model (supplemental Fig. 5). Interestingly, we also get significant REHH values at all three BEHs but the haplotypes that contain the ancestral alleles are the ones showing evidence of selection (supplemental Fig. 6). This result is likely due to the fact that the C allele of rs1800414 occurs on the same chromosome as these haplotypes in East Asia (supplemental Fig. 7). As with our European population samples we divided the East Asians into three groups: Western China, East Asia, and Southeast Asia. We see there is strong evidence of selection for the C allele of rs1800414 in all three population groups (supplemental Fig. 8). In both Western China and Southeast Asia, the frequency of the derived allele of rs1800414 is <50%, so we were able to use the nHS test on these populations. Using the nHS test we see strong evidence of selection for the derived allele of rs1800414 in both the Western China and Southeast Asian groups (Fig. 10d, e).
Selection results at rs1800414 in East Asia. This figure shows the results of an REHH test in East Asia (a, b), an nHS test in Western China (c), and an nHS test in Southeast Asia. Again, the cyan points represent the results from 1,000 simulated populations under the neutral constant population size model. In a and b we show strong evidence of selection at the derived allele of rs1800414 (CAC) in East Asia. This result is confirmed in Western China (c) and Southeast Asia (d) where the derived allele (circled in red) is <0.50 using nHS
We saw no evidence for selection at any of the pigmentation regions in Africa or the Americas (supplemental Figs. 9 and 10).
Discussion
Distribution of blue-eye associated alleles
The frequencies of the haplotypes associated with blue eyes of the three blue-eye associated haplotypes in the OCA2 and HERC2 genes are very similar in Northwestern and Eastern Europe where all three haplotypes have their highest frequencies (Fig. 2). This also holds true for homozygotes of the blue-eye associated alleles of these haplotypes (Supplemental Fig. 11). All three blue-eye associated alleles and homozygotes of these alleles are also present in Southern Europe and Southwest Asia at lower frequencies than those found in Northwestern and Eastern Europe; however, the frequencies of the TG allele of BEH2 and its homozygotes are lower than those of the ACA allele of BEH1 and the CA allele of BEH3. Outside of Europe, the blue-eye associated alleles of BEH1 and BEH2 are still common and homozygotes of these alleles are still seen but the blue-eye associated allele of BEH2 is much rarer and blue-eye associated homozygotes are virtually unseen.
Given the strong LD in Europe across all three haplotype systems, their association with the blue eye phenotype in Europe is understandable. However, these frequency data for other populations around the world and the essential restriction of blue eyes to Europe, shows that the BEH1 and BEH3 haplotype systems, and the composing SNPs are not universal markers of blue eyes. The TG allele at BEH2 is the best marker for blue eyes and may even contain the causal allele though the actual causative variant could be anywhere in the region of strong LD seen in European populations.
Global distribution of the light skin allele
We have shown that the C allele of the missense SNP rs1800414 is found almost exclusively in East Asia (Fig. 4). Within East Asia there is a general cline in the frequency of the C allele with the lowest frequencies in Western China, midrange frequencies in Southeast Asia, and high frequencies in Eastern East Asia. The major exception to this pattern is the Malaysians; in our small sample the derived allele is absent, but the Malays are an Austronesian group and they show similar frequencies to our other Austronesian populations (Micronesians and Samoans).
Selection in the OCA2-HERC2 region
We showed that the strongest signal of selection in Europe and Southwest Asia is at the TG allele of BEH2 and any signal seen at BEH1 and BEH3 is likely due to hitchhiking (Figs. 8, 9). Along with the distribution data, this strongly suggests that the TG allele of BEH2 is, contains, or is in strong LD with the blue eye causal mutation. It is possible that BEH2 is in the promoter region of OCA2 and the blue eye allele lowers the amount of OCA2 expressed either in the iris or globally.
This result also raises the question of why blue eyes would be under selection. Since there is no known biological advantage to having blue eyes, we think a likely answer is sexual selection that in Europe and Southwest Asia individuals with blue eyes are, or were, preferred as mates. Another possible explanation is that the blue eye phenotype is not being selected for; rather the TG allele of BEH2 has another phenotype, such as lighter skin pigmentation, which is under selection.
In East Asia, we show that the C allele of the missense SNP rs1800414 is also under selection (Fig. 10). Again this result is not completely unexpected since this allele has been associated with lighter skin pigmentation in East Asians, and variants affecting skin pigmentation have previously been shown to be targets of selection (Edwards et al. 2010; Izagirre et al. 2006; Lao et al. 2007; Norton et al. 2007).
Conclusions
We have shown that the TG allele of BEH2 has a much more restricted global distribution compared to the ACA allele of BEH1 and the CA allele of BEH3, the other two haplotypes published as associated with blue eyes (Duffy et al. 2007; Sturm et al. 2008; Kayser et al. 2008; Sulem et al. 2007;Mengel-From et al. 2010; Walsh et al. 2010). We also show that the TG allele of BEH2 has a strong signal of selection. Cook et al. (2009) showed melanocytes homozygous for the blue-eye associated allele of rs12913832 of BEH2 produced significantly less melanin than heterozygotes or those that were homozygous for the ancestral allele, but did not control for other SNPs in the region. This evidence suggests that BEH2 may contain the causal allele for blue eyes or at minimum is the best marker for the region in LD that does contain the causal allele. We have also shown that the C allele of rs1800414 is both restricted to East Asia and under selection in that region. This research provides further evidence for lighter pigmentation evolving by means of selection at least partly independently in Europeans and East Asians but at some genes in common.
These results, taken together with those from several forensic studies predicting iris pigmentation in mixed populations (Mengel-From et al. 2010; Spichenok et al. 2010; Valenzuela et al. 2010; Walsh et al. 2010; Pospiech et al. 2011), suggest that the SNPs of BEH2 (rs1129038 and rs12913832) are the best markers for blue eyes for forensic purposes. A recent study by Liu et al. (2010) found that rs12913832 has the strongest effect when eye color is measured quantitatively and can explain most of the variance in eye color amongst Europeans. However, several questions need to be answered. Are the SNPs in BEH2 responsible for the blue eye phenotype seen in Europeans or simply in strong LD with the causative allele? Is BEH2 in a promoter region for OCA2? Are blue eyes under sexual selection or is the TG allele also responsible for an additional selected phenotype such as light skin pigmentation? Both Eiberg et al. (2008) and Sturm et al. (2008) suggest that the BEH2 falls into a regulatory region of OCA2; however, Eiberg et al. believe the causal allele is a 166 kb haplotype that happens to contain the two SNPs of BEH2 and Sturm et al. suggest that rs12913832 is the causal allele. Eiberg et al. based their conclusion on lower activity when they used their blue-eye associated haplotype in a luciferase assay compared to other haplotypes. Sturm et al. based their conclusion on not finding a better associated SNP of known SNPs in the 5′ region of OCA2 or the 3′ end of HERC2 and that the probability of there being an unknown SNP with a stronger association was unlikely. Further research will be needed to answer these questions.
Web Resources
The URLs for data presented herein are as follows: ALFRED, http://alfred.med.yale.edu/alfred/index.asp. The International HapMap Project, http://hapmap.org/. Online Mendelian Inheritance in Man, http://www.ncbi.nlm.nih.gov/Omim.
References
Anderson MA, Gusella JF (1984) Use of cyclosporin A in establishing Epstein–Barr virus-transformed human lymphoblastoid cell lines. In Vitro 20:856–858
Anno S, Abe T, Yamamoto T (2008) Interactions between SNP alleles at multiple loci contribute to skin color differences between Caucasoid and Mongoloid subjects. Int J Biol Sci 4:81–86
Branicki W, Brudnik U, Wojas-Pelc A (2009) Interactions between HERC2, OCA2 and MC1R may influence human pigmentation phenotype. Ann Hum Genet 73:160–170
Cook AL, Chen W, Thurber AE, Smit DJ, Smith AG, Bladen TG, Brown DL, Duffy DL, Pastorino L, Bianchi-Scarra G et al (2009) Analysis of cultured human melanocytes based on polymorphisms within SLC45A2/MATP, SLC24A5/NCKX5, and OCA2/P loci. J Invest Dermatol 129:392–405
Duffy DL, Montgomery GW, Chen W, Zhao ZZ, Le L, James MR, Hayward NK, Martin NG, Sturm RA (2007) A three-single-nucleotide polymorphism haplotype in intron 1 of OCA2 explains most human eye-color variation. Am J Hum Genet 80:241–252
Edwards M, Bigham A, Tan J, Li S, Gozdzik A, Ross K, Jin L, Parra EJ (2010) Association of the OCA2 polymorphism His 615Arg with melanin content in East Asian populations: further evidence of convergent evolution of skin pigmentation. PLoS Genet 3:e1000897
Eiberg H, Troelsen J, Nielsen M, Mikkelsen A, Mengel-From J, Kjaer KW, Hansen L (2008) Blue eye color in human may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum Genet 123:177–187
Frudakis T, Thomas M, Gaskin Z, Venkateswarlu K, Chandra KS, Ginjupalli S, Gunturi S, Natrajan S, Ponnuswamy VK, Ponnuswamy KN (2003) Sequences associated with human iris pigmentation. Genetics 156:2071–2083
Gu S, Pakstis AJ, Kidd KK (2005) HAPLOT: a graphical comparison of haplotype blocks, tagSNP sets and SNP variation for multiple populations. Bioinformatics 21:3938–3939
Han Y, Gu S, Oota H, Osier M, Pakstis AJ, Speed WC, Kidd JR, Kidd KK (2007) Evidence of positive selection on a class I ADH locus. Am J Hum Genet 80:441–456
Hanchard NA, Rockett KA, Spencer C, Coop G, Pinder M, Jallow M, Kimber M, McVean G, Mott R, Kwiatkowski DP (2006) Screening for recently selected alleles by analysis of human haplotype similarity. Am J Hum Genet 78:153–159
Hudson RR (2002) Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 18:337–338
Izagirre N, Garcia I, Junquera C, de la Rua C, Alonso S (2006) A scan for signatures of positive selection in candidate loci for skin pigmentation in humans. Mol Biol Evol 23:1697–1706
Jakobsson M, Scholz SW, Scheet P, Gibbs JR, Van Liere JM, Fung HC, Szpiech ZA, Degnan JH, Wang K, Guerreiro R et al (2008) Genotype, haplotype, and copy-number variation in worldwide human populations. Nature 451:998–1003
Kayser M, Liu F, Janssens CJW, Rivadeneira F, Lao O, van Duijn K, Vermeulen M, Arp P, Jhamai MM, van IJcken WFJ et al (2008) Three genome-wide association studies and a linkage analysis identify HERC2 as a human iris color gene. Am J Hum Genet 82:411–423
Lao O, de Gruijter JM, van Duijn K, Navarro A, Kayser M (2007) Signatures of positive selection in genes associated with human skin pigmentation as revealed from analyses of single nucleotide polymorphisms. Ann Hum Genet 71:354–369
Li H, Gu S, Cai X, Speed WC, Pakstis AJ, Golub EI, Kidd JR, Kidd KK (2008) Ethnic related selection for an ADH class I variant with East Asia. PLoS ONE 3
Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL et al (2008b) Worldwide human relationships inferred from genome-wide patterns of variation. Science 319:1100–1104
Liu F, Wollstein A, Hysi PG, Ankra-Badu GA, Spector TD, Park D, Zhu G, Larsson M, Duffy DL, Montgomery GW et al (2010) Digital quantification of human eye color highlights genetic association of three new loci. PLoS Genet 6:e1000934
Mengel-From J, Borsting C, Sanchez JJ, Eiberg H, Morling N (2010) Human eye colour and HERC2, OCA2, and MATP. Forensic Sci Int Genet 4:323–328
Norton HL, Kittles RA, Parra E, McKeigue P, Mao X, Cheng K, Canfield VA, Bradley DG, McEvoy B, Shriver MD (2007) Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol Biol Evol 24:710–722
Pospiech E, Draus-Barini J, Kupiec T, Wojas-Pelc A, Branicki W (2011) Gene–gene interactions contribute to eye colour variation in humans. J Hum Genet 56:447–455
Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES (2006) Positive natural positive selection in the human lineage. Science 312:1614–1620
Scheet P, Stephens M (2006) A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet 78:629–644
Spichenok O, Budimlija ZM, Mitchell AA, Jenny A, Kovacevic L, Marjanovic D, Caragine T, Prinz M, Wurmbach E (2010) Prediction of eye and skin color in diverse populations using seven SNPs. Forensic Sci Int 5:472–478
Staleva L, Manga P, Orlow SJ (2002) Pink-eyed dilution protein modulates arsenic sensitivity and intracellular glutathione metabolism. Mol Biol Cell 13:4206–4220
Sturm RA (2009) Molecular genetics of human pigmentation diversity. Hum Mol Genet 18:R9–R17
Sturm RA, Larsson M (2009) Genetics of human iris colour and patterns. Pigment Cell Melanoma Res 22:544–562
Sturm RA, Teasdale RD, Box NF (2001) Human pigmentation genes: identification, structure and consequences of polymorphic variation. Gene 277:49–62
Sturm RA, Duffy DL, Zhao ZZ, Leite FPN, Stark MS, Hayward NK, Martin NG, Montgomery GW (2008) A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am J Hum Genet 82:424–431
Sulem P, Gudbjartsson DF, Stacey SN, Hegason A, Rafnar T, Magnusson KP, Manolescu A, Karason A, Palsson A, Thorleifsson G (2007) Genetic determinants of hair, eye, and skin pigmentation in Europeans. Nat Genet 39:1443–1452
Toyofuku K, Valencia JC, Kushimoto T, Costin GE, Virador VM, Vieira WD, Ferrans VJ, Hearing VJ (2002) The etiology of oculocutaneous albinism (OCA) type II: the pink protein modulates the processing and transport of tyrosinase. Pigment Cell Res 15:217–224
Valenzuela RK, Henderson MS, Walsh MH, Garrison NA, Kelch JT, Cohen-Barak O, Erickson DT, Meaney FJ, Walsh JB, Cheng KC et al (2010) Predicting phenotype from genotype: normal pigmentation. J Forensic Sci 55:315–322
Walsh S, Lindenbergh A, Zuniga SB, Sijen T, de Knijff P, Kayser M, Ballantyne KN (2010) Developmental validation of the IrisPlex system: determination of blue and brown iris colour for forensic intelligence. Forensic Sci Int Genet 5:467–471
Yuasa I, Umetsu K, Harihara S, Kido A, Miyoshi A, Saitou N, Dashnyam B, Jin F, Lucotte G, Chattopadhyay PK et al (2007) Distribution of two Asian-related coding SNPs in the MC1R and OCA2 genes. Biochem Genet 45:535–542
Acknowledgments
This research was funded in part by National Institutes of Health Grant GM57672 and National Institute of Justice, Office of Justice Programs, US Department of Justice Grants 2007-DN-BX-K197, 2010-DN-BX-K225 awarded to KKK. Points of view in this document are those of the authors and do not necessarily represent the official position or policies of the US Department of Justice. We would like to acknowledge all our collaborators who helped collect the samples used in this research as well as the National Laboratory for the Genetics of Israeli Populations at Tel Aviv University and the Coriell Cell Repositories. Finally we would like to thank the thousands of individuals who donated samples without whom this research would not be possible.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Donnelly, M.P., Paschou, P., Grigorenko, E. et al. A global view of the OCA2-HERC2 region and pigmentation. Hum Genet 131, 683–696 (2012). https://doi.org/10.1007/s00439-011-1110-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-011-1110-x