Abstract
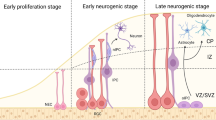
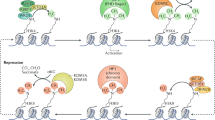
Post-translational modification of histone proteins, such as the methylation of lysine and arginine residues, influences the higher order of chromatin and leads to gene activation or silencing. Histone methyltransferases or demethylases actively add or remove various methylation marks in a cell-type-specific and context-dependent way. They are therefore important players in regulating the transcriptional program of a cell. Some control of the various cellular programs is necessary during the differentiation of stem cells along a specific lineage, when differentiation to alternative lineages needs to be suppressed. One example is the development of neurons from neural stem cells during neurogenesis. Neurogenesis is a highly organized process that requires the proper coordination of survival, proliferation, differentiation and migration signals. This holds true for both embryonic and neural stem cells that give rise to the various cell types of the central nervous system. The control of embryonic and neural stem cell self-renewal and differentiation is achieved by both extrinsic and intrinsic signals that regulate gene expression precisely. Recent advances in neuroscience support the importance of epigenetic modifications, such as the methylation and acetylation of histones, as an important intrinsic mechanism for the regulation of central nervous system development. This review summarizes our current knowledge of histone methylation processes during neural development and provides insights into the function of histone methylation enzymes and their role during central nervous system development.







Similar content being viewed by others
Abbreviations
- 5hmC:
-
5-Hydroxymethylcytosine
- CNS:
-
Central nervous system
- COMPASS:
-
Complex of proteins associated with Set1
- DOT1L:
-
Disruptor of telomeric silencing 1-like
- ESC:
-
Embryonic stem cells
- EZH2:
-
Enhancer of zeste homolog 2
- H3:
-
Histone 3
- H3K4 (same scheme for other modifications):
-
Histone 3 lysine 4
- H4R3me2a/s:
-
Histone 4 arginine 3 asymmetric/symmetric dimethylation
- HDAC:
-
Histone deacetylase
- K:
-
Lysine
- KDM:
-
Histone lysine demethylase
- KMT:
-
Histone lysine methyltransferase
- KO:
-
Knockout
- JARID:
-
Jumonji/ARID domain
- JHDM:
-
JmjC domain-containing histone demethylase
- JmjC:
-
Jumonji C
- me:
-
Methylation
- me1/2/3:
-
Mono- /di- /trimethylation
- MLL:
-
Mixed-lineage leukemia
- NSC:
-
Neural stem cells
- NTD:
-
Neural tube defects
- PRC1:
-
Polycomb repressive complex 1
- PRC2:
-
Polycomb repressive complex 2
- R:
-
Arginine
- RE:
-
Neuron-restrictive silencer element
- REST:
-
RE1-silencing transcription factor
- RNAPII:
-
RNA polymerase II
- SAH:
-
S-adenosyl-homocysteine
- SAM:
-
S-adenosyl-L-methionine
- SET:
-
Su(var)3-9, Enhancer of Zeste, Trithorax
- TSS:
-
Transcription start site
- Trx:
-
Trithorax
- Trr:
-
Trithorax-related
References
Adegbola A, Gao H, Sommer S, Browning M (2008) A novel mutation in JARID1C/SMCX in a patient with autism spectrum disorder (ASD). Am J Med Genet A 146A:505–511. doi:10.1002/ajmg.a.32142
Albert M, Schmitz SU, Kooistra SM, Malatesta M, Morales Torres C, Rekling JC, Johansen JV, Abarrategui I, Helin K (2013) The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3. PLoS Genet 9:e1003461. doi:10.1371/journal.pgen.1003461
Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG (2006) Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8:532–538. doi:10.1038/ncb1403
Barrand S, Andersen IS, Collas P (2010) Promoter-exon relationship of H3 lysine 9, 27, 36 and 79 methylation on pluripotency-associated genes. Biochem Biophys Res Commun 401:611–617. doi:10.1016/j.bbrc.2010.09.116
Barry ER, Krueger W, Jakuba CM, Veilleux E, Ambrosi DJ, Nelson CE, Rasmussen TP (2009) ES cell cycle progression and differentiation require the action of the histone methyltransferase Dot1L. Stem Cells Dayt Ohio 27:1538–1547. doi:10.1002/stem.86
Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–326. doi:10.1016/j.cell.2006.02.041
Bernstein BE, Meissner A, Lander ES (2007) The mammalian epigenome. Cell 128:669–681. doi:10.1016/j.cell.2007.01.033
Bower C, D’Antoine H, Stanley FJ (2009) Neural tube defects in Australia: trends in encephaloceles and other neural tube defects before and after promotion of folic acid supplementation and voluntary food fortification. Birt Defects Res A Clin Mol Teratol 85:269–273. doi:10.1002/bdra.20536
Burgold T, Spreafico F, De Santa F, Totaro MG, Prosperini E, Natoli G, Testa G (2008) The Histone H3 Lysine 27-Specific Demethylase Jmjd3 Is Required for Neural Commitment. PLoS One 3:e3034. doi:10.1371/journal.pone.0003034
Büttner N, Johnsen SA, Kügler S, Vogel T (2010) Af9/Mllt3 interferes with Tbr1 expression through epigenetic modification of histone H3K79 during development of the cerebral cortex. Proc Natl Acad Sci U S A 107:7042–7047. doi:10.1073/pnas.0912041107
Cao R, Zhang Y (2004) The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev 14:155–164. doi:10.1016/j.gde.2004.02.001
Chang B, Chen Y, Zhao Y, Bruick RK (2007) JMJD6 is a histone arginine demethylase. Science 318:444–447. doi:10.1126/science.1145801
Cheung I, Shulha HP, Jiang Y, Matevossian A, Wang J, Weng Z, Akbarian S (2010) Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc Natl Acad Sci 107:8824–8829. doi:10.1073/pnas.1001702107
Chittka A (2010) Dynamic Distribution of Histone H4 Arginine 3 Methylation Marks in the Developing Murine Cortex. PLoS One 5:e13807. doi:10.1371/journal.pone.0013807
Chittka A, Nitarska J, Grazini U, Richardson WD (2012) Transcription Factor Positive Regulatory Domain 4 (PRDM4) Recruits Protein Arginine Methyltransferase 5 (PRMT5) to Mediate Histone Arginine Methylation and Control Neural Stem Cell Proliferation and Differentiation. J Biol Chem 287:42995–43006. doi:10.1074/jbc.M112.392746
DeCarlo D, Hadden MK (2012) Oncoepigenomics: Making histone lysine methylation count. Eur J Med Chem 56:179–194. doi:10.1016/j.ejmech.2012.08.010
De Vos D, Frederiks F, Terweij M, van Welsem T, Verzijlbergen KF, Iachina E, de Graaf EL, Altelaar AFM, Oudgenoeg G, Heck AJR, Krijgsveld J, Bakker BM, van Leeuwen F (2011) Progressive methylation of ageing histones by Dot1 functions as a timer. EMBO Rep 12:956–962. doi:10.1038/embor.2011.131
Dey BK, Stalker L, Schnerch A, Bhatia M, Taylor-Papidimitriou J, Wynder C (2008) The Histone Demethylase KDM5b/JARID1b Plays a Role in Cell Fate Decisions by Blocking Terminal Differentiation. Mol Cell Biol 28:5312–5327. doi:10.1128/MCB.00128-08
Di Meglio T, Kratochwil CF, Vilain N, Loche A, Vitobello A, Yonehara K, Hrycaj SM, Roska B, Peters AHFM, Eichmann A, Wellik D, Ducret S, Rijli FM (2013) Ezh2 Orchestrates Topographic Migration and Connectivity of Mouse Precerebellar Neurons. Science 339:204–207. doi:10.1126/science.1229326
Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG (2006) Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol 13:713–719. doi:10.1038/nsmb1128
Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473:43–49. doi:10.1038/nature09906
Fontebasso AM, Schwartzentruber J, Khuong-Quang D-A, Liu X-Y, Sturm D, Korshunov A, Jones DTW, Witt H, Kool M, Albrecht S, Fleming A, Hadjadj D, Busche S, Lepage P, Montpetit A, Staffa A, Gerges N, Zakrzewska M, Zakrzewski K, Liberski PP, Hauser P, Garami M, Klekner A, Bognar L, Zadeh G, Faury D, Pfister SM, Jabado N, Majewski J (2013) Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol (Berl) 125:659–669. doi:10.1007/s00401-013-1095-8
Fukuda T, Tokunaga A, Sakamoto R, Yoshida N (2011) Fbxl10/Kdm2b deficiency accelerates neural progenitor cell death and leads to exencephaly. Mol Cell Neurosci 46:614–624. doi:10.1016/j.mcn.2011.01.001
Gibson WT, Hood RL, Zhan SH, Bulman DE, Fejes AP, Moore R, Mungall AJ, Eydoux P, Babul-Hirji R, An J, Marra MA, Chitayat D, Boycott KM, Weaver DD, Jones SJM (2012) Mutations in EZH2 Cause Weaver Syndrome. Am J Hum Genet 90:110–118. doi:10.1016/j.ajhg.2011.11.018
Golebiewska A, Atkinson SP, Lako M, Armstrong L (2009) Epigenetic landscaping during hESC differentiation to neural cells. Stem Cells Dayt Ohio 27:1298–1308. doi:10.1002/stem.59
Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD (2010) Histone methylation regulates memory formation. J Neurosci Off J Soc Neurosci 30:3589–3599. doi:10.1523/JNEUROSCI.3732-09.2010
Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin S-G, Jiang Y, Pfeifer GP, Lu Q (2013) Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep 3:291–300. doi:10.1016/j.celrep.2013.01.011
He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y (2013) Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol 15:373–384. doi:10.1038/ncb2702
Henriquez B, Bustos FJ, Aguilar R, Becerra A, Simon F, Montecino M, van Zundert B (2013) Ezh1 and Ezh2 differentially regulate PSD-95 gene transcription in developing hippocampal neurons. Mol Cell Neurosci. doi:10.1016/j.mcn.2013.07.012
Herz H-M, Mohan M, Garruss AS, Liang K, Takahashi Y-H, Mickey K, Voets O, Verrijzer CP, Shilatifard A (2012) Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev 26:2604–2620. doi:10.1101/gad.201327.112
Hirabayashi Y, Gotoh Y (2010) Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci 11:377–388. doi:10.1038/nrn2810
Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, Koseki H, Vidal M, Gotoh Y (2009) Polycomb Limits the Neurogenic Competence of Neural Precursor Cells to Promote Astrogenic Fate Transition. Neuron 63:600–613. doi:10.1016/j.neuron.2009.08.021
Huang S (2005) Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev 19:1885–1893. doi:10.1101/gad.1333905
Huang X, Dixit VM (2011) Cross talk between ubiquitination and demethylation. Mol Cell Biol 31:3682–3683. doi:10.1128/MCB.06001-11
Ichi S, Costa FF, Bischof JM, Nakazaki H, Shen Y-W, Boshnjaku V, Sharma S, Mania-Farnell B, McLone DG, Tomita T, Soares MB, Mayanil CSK (2010) Folic Acid Remodels Chromatin on Hes1 and Neurog2 Promoters during Caudal Neural Tube Development. J Biol Chem 285:36922–36932. doi:10.1074/jbc.M110.126714
Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y (2007) The X-Linked Mental Retardation Gene SMCX/JARID1C Defines a Family of Histone H3 Lysine 4 Demethylases. Cell 128:1077–1088. doi:10.1016/j.cell.2007.02.017
Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim H-J, Glass CK, Hermanson O, Rosenfeld MG (2007) SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 450:415–419. doi:10.1038/nature06270
Jiang H, Shukla A, Wang X, Chen W, Bernstein BE, Roeder RG (2011) Role for Dpy-30 in ES Cell-Fate Specification by Regulation of H3K4 Methylation within Bivalent Domains. Cell 144:513–525. doi:10.1016/j.cell.2011.01.020
Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, Baltus GA, Kadam S, Zhai H, Valdez R, Gonzalo S, Zhang Y, Li E, Chen T (2008) The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet 4:e1000190. doi:10.1371/journal.pgen.1000190
Jones WD, Dafou D, McEntagart M, Woollard WJ, Elmslie FV, Holder-Espinasse M, Irving M, Saggar AK, Smithson S, Trembath RC, Deshpande C, Simpson MA (2012) De Novo Mutations in MLL Cause Wiedemann-Steiner Syndrome. Am J Hum Genet 91:358–364. doi:10.1016/j.ajhg.2012.06.008
Karpiuk O, Najafova Z, Kramer F, Hennion M, Galonska C, König A, Snaidero N, Vogel T, Shchebet A, Begus-Nahrmann Y, Kassem M, Simons M, Shcherbata H, Beissbarth T, Johnsen SA (2012) The histone H2B monoubiquitination regulatory pathway is required for differentiation of multipotent stem cells. Mol Cell 46:705–713. doi:10.1016/j.molcel.2012.05.022
Kerimoglu C, Agis-Balboa RC, Kranz A, Stilling R, Bahari-Javan S, Benito-Garagorri E, Halder R, Burkhardt S, Stewart AF, Fischer A (2013) Histone-Methyltransferase MLL2 (KMT2B) Is Required for Memory Formation in Mice. J Neurosci 33:3452–3464. doi:10.1523/JNEUROSCI.3356-12.2013
Kim J, Hake SB, Roeder RG (2005) The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell 20:759–770. doi:10.1016/j.molcel.2005.11.012
Kim S-K, Jung I, Lee H, Kang K, Kim M, Jeong K, Kwon CS, Han Y-M, Kim YS, Kim D, Lee D (2012) Human histone H3K79 methyltransferase DOT1L protein [corrected] binds actively transcribing RNA polymerase II to regulate gene expression. J Biol Chem 287:39698–39709. doi:10.1074/jbc.M112.384057
Kleefstra T, Brunner HG, Amiel J, Oudakker AR, Nillesen WM, Magee A, Geneviève D, Cormier-Daire V, Van Esch H, Fryns J-P (2006) Loss-of-Function Mutations in <i> Euchromatin Histone Methyl Transferase 1</i > (<i > EHMT1</i>) Cause the 9q34 Subtelomeric Deletion Syndrome. Am J Hum Genet 79:370–377
Kleefstra T, Kramer JM, Neveling K, Willemsen MH, Koemans TS, Vissers LELM, Wissink-Lindhout W, Fenckova M, van den Akker WMR, Kasri NN, Nillesen WM, Prescott T, Clark RD, Devriendt K, van Reeuwijk J, de Brouwer APM, Gilissen C, Zhou H, Brunner HG, Veltman JA, Schenck A, van Bokhoven H (2012) Disruption of an EHMT1-Associated Chromatin-Modification Module Causes Intellectual Disability. Am J Hum Genet 91:73–82. doi:10.1016/j.ajhg.2012.05.003
Klose RJ, Kallin EM, Zhang Y (2006) JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7:715–727. doi:10.1038/nrg1945
Kouzarides T (2007) Chromatin modifications and their function. Cell 128:693–705. doi:10.1016/j.cell.2007.02.005
Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T (1997) Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J 16:3219–3232. doi:10.1093/emboj/16.11.3219
Laumonnier F (2005) Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J Med Genet 42:780–786. doi:10.1136/jmg.2004.029439
Lee BM, Mahadevan LC (2009) Stability of histone modifications across mammalian genomes: implications for “epigenetic” marking. J Cell Biochem 108:22–34. doi:10.1002/jcb.22250
Lee ER, Murdoch FE, Fritsch MK (2007a) High histone acetylation and decreased polycomb repressive complex 2 member levels regulate gene specific transcriptional changes during early embryonic stem cell differentiation induced by retinoic acid. Stem Cells Dayt Ohio 25:2191–2199. doi:10.1634/stemcells.2007-0203
Lee MG, Villa R, Trojer P, Norman J, Yan K-P, Reinberg D, Di Croce L, Shiekhattar R (2007b) Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318:447–450. doi:10.1126/science.1149042
Lee S, Roeder RG, Lee JW (2009) Roles of histone H3-lysine 4 methyltransferase complexes in NR-mediated gene transcription. Prog Mol Biol Transl Sci 87:343–382. doi:10.1016/S1877-1173(09)87010-5
Li X, Hu X, Patel B, Zhou Z, Liang S, Ybarra R, Qiu Y, Felsenfeld G, Bungert J, Huang S (2010) H4R3 methylation facilitates beta-globin transcription by regulating histone acetyltransferase binding and H3 acetylation. Blood 115:2028–2037. doi:10.1182/blood-2009-07-236059
Lim DA, Huang Y-C, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A (2009) Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458:529–533. doi:10.1038/nature07726
Lubitz S, Glaser S, Schaft J, Stewart AF, Anastassiadis K (2007) Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Mol Biol Cell 18:2356–2366. doi:10.1091/mbc.E06-11-1060
Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260. doi:10.1038/38444
Martin C, Zhang Y (2005) The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6:838–849. doi:10.1038/nrm1761
McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW (2008) Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 453:812–816. doi:10.1038/nature06906
Meaney MJ, Ferguson-Smith AC (2010) Epigenetic regulation of the neural transcriptome: the meaning of the marks. Nat Neurosci 13:1313–1318. doi:10.1038/nn1110-1313
Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–560. doi:10.1038/nature06008
Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A (2001) COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A 98:12902–12907. doi:10.1073/pnas.231473398
Mohan M, Herz H-M, Takahashi Y-H, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A (2010) Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev 24:574–589. doi:10.1101/gad.1898410
Mohan M, Herz H-M, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A (2011) The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol 31:4310–4318. doi:10.1128/MCB.06092-11
Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schübeler D (2008) Lineage-Specific Polycomb Targets and De Novo DNA Methylation Define Restriction and Potential of Neuronal Progenitors. Mol Cell 30:755–766. doi:10.1016/j.molcel.2008.05.007
Mulinare J, Cordero JF, Erickson JD, Berry RJ (1988) Periconceptional use of multivitamins and the occurrence of neural tube defects. JAMA, J Am Med Assoc 260:3141–3145
Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin C-F, Stevens C, Wang L-S, Makarov V, Polak P, Yoon S, Maguire J, Crawford EL, Campbell NG, Geller ET, Valladares O, Schafer C, Liu H, Zhao T, Cai G, Lihm J, Dannenfelser R, Jabado O, Peralta Z, Nagaswamy U, Muzny D, Reid JG, Newsham I, Wu Y, Lewis L, Han Y, Voight BF, Lim E, Rossin E, Kirby A, Flannick J, Fromer M, Shakir K, Fennell T, Garimella K, Banks E, Poplin R, Gabriel S, DePristo M, Wimbish JR, Boone BE, Levy SE, Betancur C, Sunyaev S, Boerwinkle E, Buxbaum JD, Cook EH Jr, Devlin B, Gibbs RA, Roeder K, Schellenberg GD, Sutcliffe JS, Daly MJ (2012) Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485:242–245. doi:10.1038/nature11011
Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J (2010) Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet 42:790–793. doi:10.1038/ng.646
Niwa H (2007) How is pluripotency determined and maintained? Dev Camb Engl 134:635–646. doi:10.1242/dev.02787
O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, Turner EH, Stanaway IB, Vernot B, Malig M, Baker C, Reilly B, Akey JM, Borenstein E, Rieder MJ, Nickerson DA, Bernier R, Shendure J, Eichler EE (2012) Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485:246–250. doi:10.1038/nature10989
Ooga M, Inoue A, Kageyama S, Akiyama T, Nagata M, Aoki F (2008) Changes in H3K79 methylation during preimplantation development in mice. Biol Reprod 78:413–424. doi:10.1095/biolreprod.107.063453
Outchkourov NS, Muiño JM, Kaufmann K, van Ijcken WFJ, Koerkamp MJG, van Leenen D, de Graaf P, Holstege FCP, Grosveld FG, Timmers HTM (2013) Balancing of Histone H3K4 Methylation States by the Kdm5c/SMCX Histone Demethylase Modulates Promoter and Enhancer Function. Cell Rep 3:1071–1079. doi:10.1016/j.celrep.2013.02.030
Pasini D, Hansen KH, Christensen J, Agger K, Cloos PAC, Helin K (2008) Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev 22:1345–1355. doi:10.1101/gad.470008
Pereira JD, Sansom SN, Smith J, Dobenecker M-W, Tarakhovsky A, Livesey FJ (2010) Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci U S A 107:15957–15962. doi:10.1073/pnas.1002530107
Peters AHFM, Mermoud JE, O’Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T (2002) Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet 30:77–80. doi:10.1038/ng789
Rai K, Jafri IF, Chidester S, James SR, Karpf AR, Cairns BR, Jones DA (2009) Dnmt3 and G9a Cooperate for Tissue-specific Development in Zebrafish. J Biol Chem 285:4110–4121. doi:10.1074/jbc.M109.073676
Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF (2004) Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell 14:727–738
Sanders SL, Portoso M, Mata J, Bähler J, Allshire RC, Kouzarides T (2004) Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119:603–614. doi:10.1016/j.cell.2004.11.009
Schmitz SU, Albert M, Malatesta M, Morey L, Johansen JV, Bak M, Tommerup N, Abarrategui I, Helin K (2011) Jarid1b targets genes regulating development and is involved in neural differentiation. EMBO J 30:4586–4600
Schübeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, Bell SP, Groudine M (2004) The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 18:1263–1271. doi:10.1101/gad.1198204
Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, Johnston M, Jaspersen SL, Kobor MS, Shilatifard A (2009) Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol Cell 35:626–641. doi:10.1016/j.molcel.2009.07.017
Shanower GA, Muller M, Blanton JL, Honti V, Gyurkovics H, Schedl P (2005) Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics 169:173–184. doi:10.1534/genetics.104.033191
Sher F, Boddeke E, Copray S (2011) Ezh2 expression in astrocytes induces their dedifferentiation toward neural stem cells. Cell Reprogramming 13:1–6. doi:10.1089/cell.2010.0052
Sher F, Boddeke E, Olah M, Copray S (2012) Dynamic Changes in Ezh2 Gene Occupancy Underlie Its Involvement in Neural Stem Cell Self-Renewal and Differentiation towards Oligodendrocytes. PLoS One 7:e40399. doi:10.1371/journal.pone.0040399
Sher F, Rößler R, Brouwer N, Balasubramaniyan V, Boddeke E, Copray S (2008) Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells 26:2875–2883
Shulha HP, Cheung I, Guo Y, Akbarian S, Weng Z (2013) Coordinated Cell Type–Specific Epigenetic Remodeling in Prefrontal Cortex Begins before Birth and Continues into Early Adulthood. PLoS Genet 9:e1003433. doi:10.1371/journal.pgen.1003433
Smithells RW, Sheppard S, Schorah CJ, Seller MJ, Nevin NC, Harris R, Read AP, Fielding DW, Walker S (1981) Vitamin supplementation and neural tube defects. Lancet 2:1425
Song M-R, Ghosh A (2004) FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat Neurosci 7:229–235. doi:10.1038/nn1192
Spyropoulou A, Piperi C, Adamopoulos C, Papavassiliou AG (2012) Deregulated Chromatin Remodeling in the Pathobiology of Brain Tumors. NeuroMolecular Med 15:1–24. doi:10.1007/s12017-012-8205-y
Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim J-E, Chen J, Lazar MA, Blobel GA, Vakoc CR (2008) DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol 28:2825–2839. doi:10.1128/MCB.02076-07
Sun G, Alzayady K, Stewart R, Ye P, Yang S, Li W, Shi Y (2010) Histone Demethylase LSD1 Regulates Neural Stem Cell Proliferation. Mol Cell Biol 30:1997–2005. doi:10.1128/MCB.01116-09
Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y (2007) The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 447:601–605. doi:10.1038/nature05823
Tan S-L, Nishi M, Ohtsuka T, Matsui T, Takemoto K, Kamio-Miura A, Aburatani H, Shinkai Y, Kageyama R (2012) Essential roles of the histone methyltransferase ESET in the epigenetic control of neural progenitor cells during development. Development 139:3806–3816. doi:10.1242/dev.082198
Tatton-Brown K, Hanks S, Ruark E, Zachariou A, Duarte SDV, Ramsay E, Snape K, Murray A, Perdeaux ER, Seal S (2011) Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget 2:1127–1133
Tatum D, Li S (2011) Evidence that the histone methyltransferase Dot1 mediates global genomic repair by methylating histone H3 on lysine 79. J Biol Chem 286:17530–17535. doi:10.1074/jbc.M111.241570
Tsukada Y, Ishitani T, Nakayama KI (2010) KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev 24:432–437. doi:10.1101/gad.1864410
Van Leeuwen F, Gafken PR, Gottschling DE (2002) Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109:745–756
Vogel T, Stoykova A, Gruss P (2006) Differential expression of polycomb repression complex 1 (PRC1) members in the developing mouse brain reveals multiple complexes. Dev Dyn 235:2574–2585. doi:10.3758/BF03194004
Wagner EJ, Carpenter PB (2012) Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 13:115–126. doi:10.1038/nrm3274
Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, Ge K, Krumlauf R, Shilatifard A (2009) Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol 29:6074–6085. doi:10.1128/MCB.00924-09
Xu X, Hoang S, Mayo MW, Bekiranov S (2010) Application of machine learning methods to histone methylation ChIP-Seq data reveals H4R3me2 globally represses gene expression. BMC Bioinformatics 11:396. doi:10.1186/1471-2105-11-396
Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, Evans CP, Rosenfeld MG (2013) lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 500:598–602. doi:10.1038/nature12451
Zhang Q, Xue P, Li H, Bao Y, Wu L, Chang S, Niu B, Yang F, Zhang T (2013) Histone modification mapping in human brain reveals aberrant expression of histone H3 lysine 79 dimethylation in neural tube defects. Neurobiol Dis 54:404–413. doi:10.1016/j.nbd.2013.01.014
Zhang X, Wen H, Shi X (2011) Lysine methylation: beyond histones. Acta Biochim Biophys Sin 44:14–27. doi:10.1093/abbs/gmr100
Zhang Y, Reinberg D (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15:2343–2360. doi:10.1101/gad.927301
Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, Cunningham JM, Jane SM (2009) PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol 16:304–311. doi:10.1038/nsmb.1568
Zhou VW, Goren A, Bernstein BE (2011) Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12:7–18. doi:10.1038/nrg2905
Zhu B, Zheng Y, Pham A-D, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D (2005) Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell 20:601–611. doi:10.1016/j.molcel.2005.09.025
Acknowledgement
The authors thank P.P. Bovio and S.C. Weise for graphical assistance, Dr. C.J. Hindley for language editing and the reviewers and editors for critical comments and fruitful discussions that have helped to improve the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
The authors are grateful to the grantholder, Prof. T. Vogel and the SFB992 Medical Epigenetics “MEDEP” (Deutsche Forschungsgemeinschaft) for funding our research.
Rights and permissions
About this article
Cite this article
Roidl, D., Hacker, C. Histone methylation during neural development. Cell Tissue Res 356, 539–552 (2014). https://doi.org/10.1007/s00441-014-1842-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-014-1842-8