Abstract
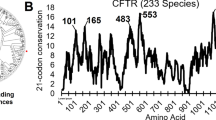
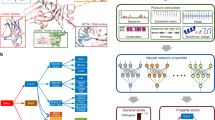
Protein modeling and molecular dynamics hold a unique toolset to aide in the characterization of clinical variants that may result in disease. Not only do these techniques offer the ability to study under characterized proteins, but they do this with the speed that is needed for time-sensitive clinical cases. In this paper we retrospectively study a clinical variant in the XIAP protein, C203Y, while addressing additional variants seen in patients with similar gastrointestinal phenotypes as the C203Y mutation. In agreement with the clinical tests performed on the C203Y patient, protein modeling and molecular dynamics suggest that direct interactions with RIPK2 and Caspase3 are altered by the C203Y mutation and subsequent loss of Zn coordination in the second BIR domain of XIAP. Interestingly, the variant does not appear to alter interactions with SMAC, resulting in further damage to the caspase and NOD2 pathways. To expand the computational strategy designed when studying XIAP, we have applied the molecular modeling tools to a list of 140 variants seen in CFTR associated with cystic fibrosis, and a list of undiagnosed variants in 17 different genes. This paper shows the exciting applications of molecular modeling in the classification and characterization of genetic variants identified in next generation sequencing.

XIAP in Caspase 3 and NOD2 signaling pathways









Similar content being viewed by others
References
Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38:W529–W533
Blom N, Gammeltoft S, Brunak S (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294:1351–1362
Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S (2004) Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4:1633–1649
Chai JJ, Du CY, Wu JW, Kyin S, Wang XD, Shi YG (2000) Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature 406:855–862
Damgaard RB, Fiil BK, Speckmann C, Yabal M, zur Stadt U, Bekker-Jensen S, Jost PJ, Ehl S, Mailand N, Gyrd-Hansen M (2013) Disease-causing mutations in the XIAP BIR2 domain impair NOD2-dependent immune signalling. EMBO Mol Med 5:1278–1295
Dinkel H, Michael S, Weatheritt RJ, Davey NE, Van Roey K, Altenberg B, Toedt G, Uyar B, Seiler M, Budd A et al (2012) ELM--the database of eukaryotic linear motifs. Nucleic Acids Res 40:D242–D251
Di Domenico T, Walsh I, Martin AJM, Tosatto SCE (2012) MobiDB: a comprehensive database of intrinsic protein disorder annotations. Bioinforma Oxf Engl 28:2080–2081
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T et al (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24:1999–2012
Eckelman BP, Salvesen GS, Scott FL (2006) Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep 7:988–994
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783
Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A et al (2011) COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 39:D945–D950
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C et al (2013) STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41:D808–D815
Huang X, Wu Z, Mei Y, Wu M (2013) XIAP inhibits autophagy via XIAP-Mdm2-p53 signalling. EMBO J 32:2204–2216
Jacob HJ, Abrams K, Bick DP, Brodie K, Dimmock DP, Farrell M, Geurts J, Harris J, Helbling D, Joers BJ et al (2013) Genomics in clinical practice: lessons from the front lines. Sci Transl Med 5:194cm5
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci CABIOS 8:275–282
Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM (2006) MUSTANG: a multiple structural alignment algorithm. Proteins 64:559–574
Krieg A, Correa RG, Garrison JB, Le Negrate G, Welsh K, Huang Z, Knoefel WT, Reed JC (2009) XIAP mediates NOD signaling via interaction with RIP2. Proc Natl Acad Sci USA 106:14524–14529
Krieger E, Koraimann G, Vriend G (2002) Increasing the precision of comparative models with YASARA NOVA—a self-parameterizing force field. Proteins 47:393–402
Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K (2009) Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins 77(Suppl 9):114–122
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lin Y-F, Lai T-C, Chang C-K, Chen C-L, Huang M-S, Yang C-J, Liu H-G, Dong J-J, Chou Y-A, Teng K-H et al (2013) Targeting the XIAP/caspase-7 complex selectively kills caspase-3-deficient malignancies. J Clin Invest 123:3861–3875
Liu Z, Cao J, Ma Q, Gao X, Ren J, Xue Y (2011) GPS-YNO2: computational prediction of tyrosine nitration sites in proteins. Mol Biosyst 7:1197–1204
Mahadevan D, Chalasani P, Rensvold D, Kurtin S, Pretzinger C, Jolivet J, Ramanathan RK, Von Hoff DD, Weiss GJ (2013) Phase I trial of AEG35156 an antisense oligonucleotide to XIAP plus gemcitabine in patients with metastatic pancreatic ductal adenocarcinoma. Am J Clin Oncol 36:239–243
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791
Muse SV, Gaut BS (1994) A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol 11:715–724
Prokop JW, Santos RAS, Milsted A (2013) Differential mechanisms of activation of the Ang peptide receptors AT1, AT2, and MAS: using in silico techniques to differentiate the three receptors. PLoS One 8:e65307
Prokop JW, Petri V, Shimoyama ME, Watanabe IKM, Casarini DE, Leeper TC, Bilinovich SM, Jacob HJ, Santos RAS, Martins AS et al (2015) Structural libraries of protein models for multiple species to understand evolution of the renin-angiotensin system. Gen Comp Endocrinol 215:106–116
Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, Goebl MG, Iakoucheva LM (2010) Identification, analysis, and prediction of protein ubiquitination sites. Proteins 78:365–380
Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X (2008) CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng Des Sel PEDS 21:639–644
Rigaud S, Fondanèche M-C, Lambert N, Pasquier B, Mateo V, Soulas P, Galicier L, Le Deist F, Rieux-Laucat F, Revy P et al (2006) XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature 444:110–114
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738
Saleem M, Qadir MI, Perveen N, Ahmad B, Saleem U, Irshad T, Ahmad B (2013) Inhibitors of apoptotic proteins: new targets for anticancer therapy. Chem Biol Drug Des 82:243–251
Shi YG (2002) Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell 9:459–470
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539
Speckmann C, Ehl S (2014) XIAP deficiency is a mendelian cause of late-onset IBD. Gut 63:1031–1032
Speckmann C, Lehmberg K, Albert MH, Damgaard RB, Fritsch M, Gyrd-Hansen M, Rensing-Ehl A, Vraetz T, Grimbacher B, Salzer U et al (2013) X-linked inhibitor of apoptosis (XIAP) deficiency: the spectrum of presenting manifestations beyond hemophagocytic lymphohistiocytosis. Clin Immunol Orlando Fla 149:133–141
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G et al (2012) Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337:64–69
Trott O, Olson AJ (2010) AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Veillette A, Pérez-Quintero L-A, Latour S (2013) X-linked lymphoproliferative syndromes and related autosomal recessive disorders. Curr Opin Allergy Clin Immunol 13:614–622
Worthey EA, Mayer AN, Syverson GD, Helbling D, Bonacci BB, Decker B, Serpe JM, Dasu T, Tschannen MR, Veith RL et al (2011) Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med Off J Am Coll Med Genet 13:255–262
Xu D, Zhang Y (2012) Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 80:1715–1735
Xue Y, Liu Z, Gao X, Jin C, Wen L, Yao X, Ren J (2010) GPS-SNO: computational prediction of protein S-nitrosylation sites with a modified GPS algorithm. PLoS One 5:e11290
Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J (2014) GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res 42:W325–W330
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Prokop, J.W., Lazar, J., Crapitto, G. et al. Molecular modeling in the age of clinical genomics, the enterprise of the next generation. J Mol Model 23, 75 (2017). https://doi.org/10.1007/s00894-017-3258-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-017-3258-3