Abstract
Mg-chelatase H subunit (CHLH) is a multifunctional protein involved in chlorophyll synthesis, plastid-to-nucleus retrograde signaling, and ABA perception. However, whether CHLH acts as an actual ABA receptor remains controversial. Here we present evidence that CHLH affects ABA signaling in stomatal guard cells but is not itself an ABA receptor. We screened ethyl methanesulfonate-treated Arabidopsis thaliana plants with a focus on stomatal aperture-dependent water loss in detached leaves and isolated a rapid transpiration in detached leaves 1 (rtl1) mutant that we identified as a novel missense mutant of CHLH. The rtl1 and CHLH RNAi plants showed phenotypes in which stomatal movements were insensitive to ABA, while the rtl1 phenotype showed normal sensitivity to ABA with respect to seed germination and root growth. ABA-binding analyses using 3H-labeled ABA revealed that recombinant CHLH did not bind ABA, but recombinant pyrabactin resistance 1, a reliable ABA receptor used as a control, showed specific binding. Moreover, we found that the rtl1 mutant showed ABA-induced stomatal closure when a high concentration of extracellular Ca2+ was present and that a knockout mutant of Mg-chelatase I subunit (chli1) showed the same ABA-insensitive phenotype as rtl1. These results suggest that the Mg-chelatase complex as a whole affects the ABA-signaling pathway for stomatal movements.
Similar content being viewed by others
Introduction
In higher plants, the stomata, surrounded by pairs of guard cells, are the pores in the plant epidermis that regulate gas exchange between the leaves and the atmosphere. Opening of the stomata allows both transpiration and CO2 entry for photosynthesis. Under drought stress, the phytohormone abscisic acid (ABA) induces stomatal closure to prevent water loss (Schroeder et al. 2001; Shimazaki et al. 2007). ABA-induced stomatal closure is driven by an efflux of K+ from the guard cells through voltage-dependent outward-rectifying K+ channels in the plasma membranes. Activation of the K+ channels requires depolarization of the plasma membrane, and this depolarization is mainly achieved by the activation of anion channels within the plasma membrane (Schroeder et al. 1987; Kim et al. 2010).
Recently, the family of proteins containing pyrabactin resistance (PYR), pyrabactin resistance 1-like (PYL), and regulatory component of ABA receptor (RCAR) has been identified as a reliable family of ABA receptors, and ABA recognition by the PYR/PYL/RCAR family of proteins activates the SnRK2 family of protein kinases through inactivation of their central negative regulators, the type 2C protein phosphatases (PP2Cs) (Ma et al. 2009; Park et al. 2009; Santiago et al. 2009; Cutler et al. 2010). The pyr1 pyl1 pyl2 pyl4 quadruple mutant exhibited a phenotype with strong ABA-insensitive seed germination, root growth, gene expression (Park et al. 2009), and stomatal opening and closing responses (Nishimura et al. 2010), indicating a functional redundancy within the PYR/PYL/RCAR family proteins. More recently, SLAC1, which is thought to be a slow-type anion channel (Negi et al. 2008; Vahisalu et al. 2008), was shown to undergo phosphorylation via an SnRK2 family protein kinase and induce depolarization of the plasma membrane (Geiger et al. 2009; Lee et al. 2009). In addition to PYR/PYL/RCAR family proteins, several candidate ABA receptors have been reported, including the Mg-chelatase H subunit (CHLH) (Shen et al. 2006; Wu et al. 2009), G-protein coupled receptor 2 (GCR2) (Liu et al. 2007), and G-protein coupled receptor-type G proteins (GTG1 and GTG2) (Pandey et al. 2009). It should be noted that CHLH and GCR2, the ABA receptor candidates, have been controversially debated (McCourt and Creelman 2008; Cutler et al. 2010).
CHLH is one of the three subunits of Mg-chelatase (D, H, and I subunits). Mg-chelatase complex is involved in the biosynthetic pathway of chlorophyll, catalyzing the insertion of Mg2+ into protoporphyrin IX to form Mg-protoporphyrin IX (Gibson et al. 1995; Willows et al. 1996; Huang and Li 2009). CHLH has also been reported as genomes uncoupled 5 (GUN5), a regulator of plastid-to-nucleus retrograde signaling (Mochizuki et al. 2001). Furthermore, CHLH was identified as an ABA-specific binding protein in Vicia faba (Zhang et al. 2002). Subsequent extensive genetic and biochemical analyses using Arabidopsis suggested that CHLH specifically binds ABA and mediates ABA-signaling pathways involved in seed germination, root elongation, gene expression, and stomatal closure (Shen et al. 2006; Wu et al. 2009). More recently, knockout of a group of WRKY transcription factors (WRKY40, WRKY18, and WRKY60) in cch mutant has shown to rescue ABA-insensitive phenotypes of cch, including stomatal movements, seed germination and post-germination growth, suggesting that these WRKY transcription factors function as negative regulators of ABA signaling (Shang et al. 2010). The expression of CHLH is also suppressed by a key component of the circadian clock, TOC1, which interacts with the CHLH promoter; overexpression of TOC1 was shown to give rise to a phenotype with stomatal guard cells that were insensitive to ABA, as did also RNAi-mediated knockdown of CHLH (Legnaioli et al. 2009). In contrast, Müller and Hansson (2009) reported that recombinant Xan-F, an ortholog of CHLH in barley, did not bind ABA, and that xan-f loss-of-function mutants showed normal ABA responsiveness. Thus, whether CHLH functions as an ABA receptor remains controversial. Further investigations are required to elucidate the role of CHLH in ABA signaling.
Several secondary messengers regulate ABA signaling in stomatal guard cells, including Ca2+, reactive oxygen species, nitric oxide, phosphatidic acid, inositol derivatives, and sphingolipids (Kim et al. 2010). Of these, involvement of Ca2+ in ABA signaling has been well established. Cytosolic Ca2+ elevation and/or oscillation play an important role in ABA-induced stomatal closure (Allen et al. 1999, 2000, 2001; Islam et al. 2010). Treatment of the epidermis with Ca2+-chelating agents such as EGTA suppresses ABA-induced stomatal closure (Hwang and Lee 2001). These results suggest that Ca2+ acts as a signal mediator in ABA-induced stomatal closure. Moreover, the sensitivity of stomatal closing in response to elevations in the cytosolic Ca2+ concentration has been suggested to be enhanced (primed) by ABA (Young et al. 2006). A Ca2+-independent pathway in the ABA signaling of stomatal guard cells has also been reported (Allan et al. 1994; Marten et al. 2007; Siegel et al. 2009).
In the present study, we performed a screen focused on stomatal aperture-dependent transpiration in detached leaves from Arabidopsis thaliana that had been treated with ethyl methanesulfonate (EMS) to induce mutations. Consequently, we isolated a rapid transpiration in detached leaves 1 (rtl1) mutant in which the stomatal movements were insensitive to ABA, and which we identified as a novel missense mutant of the Mg-chelatase H subunit (CHLH). Phenotypic and ABA-binding analyses suggested that CHLH affects ABA signaling in stomatal movements but is not itself an ABA receptor. We propose a novel hypothesis regarding the role of CHLH in ABA signaling of stomatal guard cells.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana gl1 [Columbia (Col), carrying homozygous recessive gl1], used here as the wild type (WT), is the background ecotype of an rtl1 mutant. rtl1 was backcrossed with gl1 three times. Col is the background ecotype of a cch mutant (Mochizuki et al. 2001) and a T-DNA insertion mutant of CHLI1 (chli1; SAIL_230_D11). The transgenic line pOCA107-2 is the genetic background of a gun5-1 mutant (Mochizuki et al. 2001). The chli1 mutant was obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH, USA). Plants were grown in soil under 16-h fluorescent light (50 μmol m−2 s−1)/8-h dark cycle at 24°C in 55–70% humidity in a growth room.
To obtain the homozygous mutant line of chli1, the plant was identified by PCR using T-DNA left-border primer LB1 (5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-3′) and CHLI1 gene-specific primer (5′-GGAATCCAAATAAGGCCAAAG-3′).
Isolation of the rtl1 mutant and identification of the RTL1 locus
EMS-treated gl1 M2 seeds, purchased from Lehle Seeds (Round Rock, TX, USA), were germinated and grown under the same conditions as above. For the screening of leaf transpiration mutants, the fresh weight of a detached rosette leaf was measured at 0 and 90 min after detachment from each 4-week-old M2 plant. Individuals that showed a rapid or slow weight change compared to WT plants were selected as candidates for rapid-transpiration or slow-transpiration mutants, respectively. To determine stomatal phenotypes, we measured stomatal apertures in the isolated epidermis of 4-week-old candidate plants under several conditions using a microscope. With this screening strategy, we successfully isolated three rapid transpiration in detached leaves (rtl) mutants and two slow transpiration in detached leaves (stl) mutants.
To generate mapping populations, the rtl1 mutant was crossed with the Landsberg erecta (Ler) accession of A. thaliana. The rtl1 DNA was isolated from 143 F2 plants that showed a pale-green phenotype. DNA was isolated from individual mutant plants and mapping was performed using simple-sequence length polymorphism (SSLP) markers.
Vector construction for plant transformation
A genomic DNA fragment containing the CHLH gene, including its promoter region (from −2,814 to 5,748 bp of the CHLH locus; gCHLH), from WT plants was amplified by PCR using the specific primer set 5′-CAGCAGCCACGAGTCCTGATACAGCTCG-3′ and 5′-GTCTCGTGTCACGGCTACTGCAGATGAAGATG-3′. The amplified 8,562-bp DNA was cloned into the Gateway entry vector pCR8/GW/TOPO (Invitrogen, Carlsbad, CA, USA) and recombined by the LR reaction into the binary vector pGWB1 (Nakagawa et al. 2007). The resulting pGWB1-gCHLH vector was used to transform rtl1 plants for the complementation test. RNA interference (RNAi) lines with downregulated CHLH were generated as previously reported (Shen et al. 2006), with some modifications. A gene-specific 653-bp fragment, which was located 2,363–3,015 bp downstream from the start codon, was amplified by PCR using the primer pair 5′-ACAGAGATTCTGTGGTTGGGAAAG-3′ and 5′-GGCACTTGCCATTGCTGCTG-3′. The PCR product was introduced into pCR8/GW/TOPO and transferred into the binary vector pYU501 (Ueno et al. 2007). The resulting pYU501-CHLH RNAi was then used for transformation of WT plants. Transformation was performed using the GV3101 strain of Agrobacterium tumefaciens and the floral dip method (Clough and Bent 1998).
Measurement of stomatal aperture
Stomatal apertures were measured according to Inoue et al. (2008) with some modifications. The epidermal tissues isolated from dark-adapted 4- to 6-week-old plants were incubated in basal buffer (5 mM MES-BTP, pH 6.5, 50 mM KCl, and 0.1 mM CaCl2). For inhibition of light-induced stomatal opening by ABA, the epidermal tissues were incubated under blue/red light [blue light (Stick-B-32; EYELA, Tokyo, Japan) at 10 μmol m−2 s−1 superimposed on background red light (LED-R; EYELA) at 50 μmol m−2 s−1] for 2.5 h at 24°C in the presence or absence of 20 μM ABA. For the ABA-induced stomatal closure, the pre-illuminated epidermal tissues were incubated under blue/red light for 2.5 h with or without 20 μM ABA. Stomatal apertures in the abaxial epidermis were measured microscopically. Stomatal apertures are presented as the mean of 25 stomata with standard deviation (SD). Results were confirmed by blind reassessment by another researcher.
RT-PCR
The total RNA was extracted from seedlings or leaves of 4- to 6-week-old plants using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s instructions. First-strand cDNA was synthesized using the Takara PrimeScript II 1st Strand cDNA Synthesis Kit (Takara, Tokyo, Japan) and used as a template. A 741-bp fragment of CHLH cDNA was amplified using the primer pair 5′-GTGTGAGACCAATTGCTGATAC-3′ and 5′-ACTCCATCCCACAGTGTTGG-3′. A 952-bp fragment of CHLI1 cDNA was amplified using the primer pair 5′-GGAATCCAAATAAGGCCAAAG-3′ and 5′-ACCCATCAACATTGAGCTCTG-3′. TUB2 (At5g62690), used as a control, was amplified using the primer pair 5′-CATTGTTGATCTCTAAGATCCGTG-3′ and 5′-TACTGCTGAGAACCTCTTGAG-3′.
Immunoblot
Immunoblot analysis was performed according to Hayashi et al. (2010) with modification. Seedlings or leaves from 4- to 6-week-old plants were ground in extraction buffer (50 mM MOPS–KOH, pH 7.5, 2.5 mM EDTA, 100 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 20 μM leupeptin, and 2 mM DTT) using a mortar and pestle. Fifty micrograms of protein was loaded and separated by SDS-polyacrylamide gel electrophoresis. To detect CHLH, the polyclonal antibody raised in rabbits against recombinant CHLH protein was used for immunoblot analysis. The 14-3-3 proteins were detected with the anti-14-3-3 protein (GF14ø) antibody (Kinoshita and Shimazaki 1999) as a control. The anti-CHLH and anti-14-3-3 protein antibodies were used at a 3,000-fold dilution.
Measurement of chlorophyll contents
The chlorophyll content of rosette leaves from 4-week-old plants was determined as previously described (Moran 1982).
Preparation of recombinant proteins
The coding sequences of CHLH (from 145 to 4,146 bp; At5g13630) and full-length PYR1 (At4g17870) and ABI1 (At4g26080) were amplified by PCR using cDNA prepared from WT plants. The primers used were the following: CHLH (5′-TCTGCTGTATCTGGAAACGGC-3′ and 5′-TTATCGATCGATCCCTTCGATCTTG-3′), PYR1 (5′-ATGCCTTCGGAGTTAACACC-3′ and 5′-TCACGTCACCTGAGAACCAC-3′), ABI1 (5′-ATGGAGGAAGTATCTCCGGC-3′ and 5′-TCAGTTCAAGGGTTTGCTCTTGAG-3′). The PCR products were initially cloned into pCR8/GW/TOPO and transferred into a pDEST17 destination vector containing a 6× His epitope-tag (Invitrogen) or into pDEST15 destination vector containing GST-tag (Invitrogen) by the LR reaction. Each construct was transformed into E. coli BL21 strains, and protein expression was induced by the addition of 0.1 mM isopropyl thiogalactoside, with overnight incubation at 30°C. Purification of recombinant His-tagged CHLH and PYR1 proteins was carried out using the His Bind Kit (Novagen, Madison, WI, USA) and Ni–NTA agarose (Qiagen). Purification of recombinant His-tagged ABI1 protein was performed by the same method as CHLH or PYR1, except that all buffers contained 5 mM MgCl2 (Melcher et al. 2009). The purified CHLH protein was used for the ABA-binding assay and as an antigen for preparing the anti-CHLH antibody. The recombinant GST-CHLH and GST-PYR1 proteins were purified with glutathione Sepharose 4B beads (GE Healthcare, Uppsala, Sweden). Protein concentrations were determined with the Bio-Rad protein assay kit using bovine serum albumin as a standard.
ABA-binding assay
ABA binding to purified E. coli-expressed recombinant CHLH protein was assayed using 3H-labeled (±)-ABA (370 MBq μmol−1; American Radiolabeled Chemicals, Inc., St. Louis, MO, USA) by both the filter and pull-down methods.
For the filter method (Melcher et al. 2009; Wu et al. 2009), 2 μM purified His-tagged CHLH, PYR1, and ABI1 proteins and 50 nM 3H-labeled ABA were incubated in 0.2 mL binding buffer (10 mM Tris-Mes, pH 7.0, 2 mM MgCl2, 1 mM CaCl2, 1 mM DTT, and 250 mM mannitol) with or without 1,000-fold unlabeled ABA (no. A1049; Sigma, St. Louis, MO, USA) for 1 h at 25°C. The 3H-labeled ABA-bound protein was separated from free 3H-labeled ABA by filtering using GF/F glass fiber filter (Whatman, Little Chalfont, Buckinghamshire, UK) and washed with 5 mL ice-cold binding buffer three times. Then, the 3H-labeled ABA remaining on the filter was quantified using a liquid scintillation counter (LSC-5100; Aloka, Tokyo, Japan). PYR1 with ABI1 was used as a control of ABA binding.
For the pull-down method (Melcher et al. 2009), GST-CHLH (30 μg) or GST-PYR1 (4.3 μg) protein-bound glutathione Sepharose 4B beads and 50 nM 3H-labeled ABA were incubated in 0.2 mL binding buffer with or without 1,000-fold unlabeled ABA for 1 h at 25°C. For the binding assay of GST-PYR1, the reaction mixture were supplemented with the purified His-ABI1 (9.5 μg). Then, the beads were washed three times with the binding buffer and the radioactivity of the bound 3H-labeled ABA was measured using a liquid scintillation counter.
Seed germination and root growth assays
Seed germination and root growth assays were performed as previously reported (Shen et al. 2006). For the seed germination test, approximately 100 sterilized seeds were planted on a plate containing Murashige and Skoog inorganic salts (MS medium, pH 5.9), 3% sucrose, and 0.8% agar in the presence of 0 and 3 μM ABA. The plate was kept at 4°C for 3 days, then incubated at 24°C under fluorescent light (50 μmol m−2 s−1). The number of germinated seeds was determined at the indicated times after start of incubation. For the root growth assay, 4-day-old seedlings grown on a MS plate under fluorescent light were transferred to MS medium containing 0, 5, 10, or 20 μM ABA. After 7 days, the root length was measured.
Results
An rtl1 mutant phenotype had stomatal guard cells that were insensitive to ABA
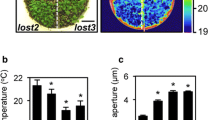
To elucidate the signaling mechanisms of stomatal opening and closing, we performed a screen focused on stomatal aperture-dependent water loss via transpiration in detached leaves using a microbalance to weigh the leaves. We selected mutants showing rapid or slow weight changes compared to the WT in detached leaves from 12,338 M2 plants treated with EMS and successfully isolated three rapid-transpiration mutants, which we designated as rtl, and two slow-transpiration mutants, which we designated as stl. Of these, rtl1 showed rapid weight changes in detached leaves under the growth conditions. The average weight of the detached rosette leaves from WT plants decreased to 65% of their initial weight over the course of 90 min, whereas the weight of the rtl1 leaves decreased to 35% of their initial weight (Fig. 1a). In addition, the rtl1 mutant was recessive and showed semi-dwarf and pale green phenotypes (Fig. 1b). The chlorophyll content of the rtl1 mutant was approximately 35% that of the WT (Fig. 2d).
Characterization of the rtl1 mutant. a Kinetics of the fresh weight change in the detached rosette leaves from 4-week-old wild-type (WT) (open circles) and rtl1 (closed circles) plants. The relative weights of leaves are presented as a percentage of the initial weight, which was the weight of each rosette leaf immediately after detachment from the plants, with the standard deviation (SD). b Four-week-old WT and rtl1 plants. Bar 10 mm. c Effect of ABA on light-induced stomatal opening in WT and rtl1 plants. Epidermal tissues from dark-adapted plants (shaded) were incubated under light (blue light at 10 μmol m−2 s−1 superimposed on background red light at 50 μmol m−2 s−1) for 2.5 h with (black) or without (white) 20 μM ABA. Data represent the mean with SD. d ABA-induced stomatal closing in WT and rtl1 plants. Pre-illuminated epidermal tissues were incubated under light (same as above) for 2.5 h with (black) or without (white) 20 μM ABA. Data represent the mean with SD. All experiments were repeated three times on different occasions with similar results
CHLH missense mutation is responsible for the rtl1 phenotype. a Mapping analysis of the RTL1 locus. The RTL1 locus was close to SSLP marker nga151 and Mg-chelatase H subunit (CHLH). b Schematic representation of the structure of the CHLH gene (upper). Black boxes and lines represent exons and introns, respectively. The position of the amino acid substitution (L690 to F) in rtl1 is indicated. The partial sequences of CHLH cDNA and the deduced amino acid in wild-type (WT) and rtl1 are shown (lower). A single nucleotide substitution (C2068 to T) is shown by a box. The amino acid and nucleotide numbers are indicated on the right. c Typical phenotypes in the WT, rtl1, and two independent gCHLH/rtl1 complementation lines (#1 and #2). Plants were grown on soil for 4 weeks. Bar 10 mm. d Chlorophyll contents in rosette leaves of 4-week-old WT, rtl1, and gCHLH/rtl1 lines (#1 and #2). The bars show the contents of chl a (white) and chl b (black). e ABA-induced stomatal closing in WT, rtl1, and gCHLH/rtl1 lines (#1 and #2). Conditions were the same as shown in Fig. 1d. Data represent the mean with SD. f ABA-induced stomatal closing in the known chlh mutants, cch and gun5. Col and pOCA are the background plants of the cch and gun5-1 mutation, respectively. Conditions were the same as shown in Fig. 1d. Data represent the mean with SD. All experiments were repeated three times on different occasions with similar results
We then analyzed the stomatal responses of rtl1 in more detail. The WT stomata closed in darkness and opened when exposed to light, and this was inhibited by 20 μM ABA. Notably, the stomata of rtl1 plants opened moderately in darkness, and light-induced stomatal opening was not inhibited by ABA (Fig. 1c). Moreover, the rtl1 plants did not show ABA-induced stomatal closure (Fig. 1d). Thus, we suspected that rapid transpiration phenotype of rtl1 is likely due to insensitivity to ABA after detachment of rosette leaves since stomatal aperture under light condition in rtl1 is almost same with that in wild type (Fig. 1c, d). In addition, seed germination and root growth showed normal ABA sensitivities in the rtl1 plants (Fig. S1). These results indicate that the ABA-insensitive phenotype of rtl1 is specific to stomatal movements.
A missense mutation of CHLH was responsible for the ABA-insensitive rtl1 phenotype
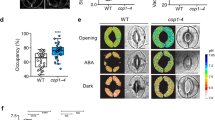
To identify the RTL1 locus, we performed a map-based analysis, which revealed that rtl1 showed a strong linkage to the SSLP marker nga151 on chromosome 5 (Fig. 2a). Our search of The Arabidopsis Information Resource (TAIR) database revealed Mg-chelatase H subunit (CHLH, At5g13630) as a candidate gene because the CHLH locus is very close to nga151, and a known CHLH missense mutant, cch (P642 to L), is pale green and ABA-insensitive (Mochizuki et al. 2001; Shen et al. 2006). We then sequenced both genomic CHLH DNA and CHLH cDNA from rtl1 and found a single nucleotide substitution from C2068 to T, which induced a novel missense mutation from L690 to F (Fig. 2b). Transformation of the WT genomic CHLH gene with its own promoter into rtl1 (gCHLH/rtl1) complemented all phenotypes that had been lost in rtl1. That is, gCHLH/rtl1 plants showed normal plant growth and chlorophyll content compared to the WT (Fig. 2c, d). Furthermore, ABA sensitivity of the stomata was restored in the gCHLH/rtl1 plants (Fig. 2e). These results indicate that the missense mutation of CHLH (L690 to F) was responsible for the rtl1 phenotype with ABA-insensitive stomatal movements. Also, the level of CHLH transcript in rosette leaves of rtl1 plants was almost identical to that of the WT, whereas the amount of CHLH protein in rosette leaves of rtl1 plants was approximately three times higher than that of the WT (Fig. 3a, b).
Phenotypes in wild-type (WT), rtl1, and CHLH RNAi lines. a RT-PCR analysis of CHLH in WT, rtl1, and two CHLH RNAi lines (#1 and #2). Total RNA was isolated from rosette leaves from 4-week-old plants. PCRs were performed with 26 cycles. TUB2 was amplified as a control. b Immunoblot analysis of CHLH protein in WT, rtl1, and CHLH RNAi lines. Fifty micrograms of protein extracted from rosette leaves was loaded on each lane. The 14-3-3 proteins were detected using anti-14-3-3 antibody as a control. c Four-week-old WT, rtl1, and CHLH RNAi plants. Bar 10 mm. d Chlorophyll content of rosette leaves from 4-week-old WT, rtl1, and CHLH RNAi plants. The bars show the content of chl a (white) and chl b (black). e Effect of ABA on stomatal closing in WT, rtl1, and CHLH RNAi plants. Conditions were the same as in Fig. 1d. Data represent the mean with SD. All experiments repeated three times on different occasions gave similar results
To confirm these results, we prepared CHLH knockdown plants. Because T-DNA insertion mutants in the CHLH gene are lethal (Shen et al. 2006), we instead prepared CHLH RNAi plants, which exhibited lower amounts of the CHLH transcript and CHLH protein (Fig. 3a, b). The CHLH RNAi plants exhibited a semi-dwarf and pale-green phenotype and did not show ABA-induced stomatal closure (Fig. 3c–e). The CHLH missense mutant, cch (P642 to L), also demonstrated ABA insensitivity, but another CHLH missense mutant, gun5-1 (A990 to V), showed normal ABA-induced stomatal closure (Fig. 2f), consistent with a previous report (Shen et al. 2006). The ABA sensitivities of seed germination and root growth were normal in both cch and rtl1 plants under our growth conditions (Fig. S1). Taken together, these results suggest that CHLH plays a role in the ABA-signaling pathway involved in stomatal movements.
Recombinant CHLH did not bind ABA
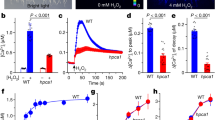
Assays of ABA binding to recombinant CHLH have implicated CHLH as an ABA receptor in Arabidopsis (Shen et al. 2006; Wu et al. 2009). On the other hand, Müller and Hansson (2009) showed that recombinant XanF, an ortholog of CHLH in barley, did not bind ABA. To clarify whether CHLH acts as an ABA receptor, we examined the ABA-binding ability of recombinant CHLH protein using 3H-labeled ABA, according to the previous conditions (Melcher et al. 2009; Wu et al. 2009). Figure 4a shows the purified recombinant CHLH (49–1,381 a.a.) truncated N-terminal plastid-transit peptide used for the ABA-binding assay. Figure 4b shows the purified reliable ABA receptor PYR1 with ABI1 as a positive control. Using the filter method, no specific ABA binding to the recombinant CHLH was detected (Fig. 4c). In contrast, PYR1 in the presence of ABI1 did bind 3H-labeled ABA, and this binding was competitively inhibited by unlabeled ABA. To confirm these results, we performed the ABA-binding assay using the pull-down method. GST-CHLH did not bind ABA, but GST-PYR1 and ABI1 specifically bound ABA (Fig. S2). These results indicate that CHLH does not bind ABA under our experimental conditions.
ABA-binding assay of the recombinant CHLH protein. a, b Coomassie Brilliant Blue staining of the recombinant His-tagged CHLH, PYR1, and ABI1 proteins used for the ABA-binding assay. Asterisks indicate degradation products of CHLH. The sizes of the molecular weight markers are shown on the left. c Determination of ABA-binding activity of the recombinant proteins by the filter method. Two micromolar recombinant protein and 50 nM 3H-labeled ABA were incubated in the absence (black) or presence of 1,000-fold unlabeled ABA (white) for 1 h at 25°C. Data represent the mean of three independent experiments with SD
Note that in our assays, we could not detect binding of ABA to PYR1 alone, and ABA binding by PYR1 required the presence of ABI1 (Fig. 4c). In accord with this finding, the ABA-binding affinity of PYL5 and RCAR1/PYL9 was reported to increase more than tenfold in the presence of PP2Cs (e.g., HAB1 and ABI2) (Ma et al. 2009; Santiago et al. 2009).
High concentration of extracellular Ca2+ restored ABA responsiveness of rtl1 stomata
Although the results of the previous section suggest that CHLH is not an ABA receptor (Fig. 4), the CHLH missense mutants, rtl1 and cch, as well as CHLH RNAi plants, all exhibited phenotypes in which stomatal movements were insensitive to ABA (Figs. 1, 2, 3). To explain these results, CHLH may have affected downstream signaling after ABA perception in the guard cells. Studies have demonstrated that Ca2+ acts as a signal mediator in ABA-induced stomatal closure (Schroeder et al. 2001, Kim et al. 2010). Thus, we examined the effect of a high extracellular concentration of Ca2+ on ABA-induced stomatal closure in the rtl1 mutant (Fig. 5). Application of 5 mM Ca2+ to the epidermis slightly reduced stomatal apertures in WT and rtl1 plants. Unexpectedly, however, ABA also induced stomatal closure in rtl1 in the presence of 5 mM Ca2+. These results suggest that a high concentration of extracellular Ca2+ restored ABA responsiveness to the rtl1 stomata.
Effect of a high concentration of Ca2+ on the ABA-induced stomatal closing in rtl1 mutant. Stomatal apertures of WT and rtl1 were measured after incubation for 2.5 h in the presence of 0.1 and 5 mM Ca2+ with or without 20 μM ABA. Other conditions were the same as in Fig. 1d. Data represent the mean with SD. Experiments repeated three times on different occasions gave similar results
A chli1 knockout mutant also had an ABA-insensitive phenotype
In addition to the H subunit, two other subunits of Mg-chelatase, the D and I subunits, are required for enzymatic activity in chlorophyll biosynthesis (Gibson et al. 1995; Willows et al. 1996; Huang and Li 2009). To investigate whether other subunits of Mg-chelatase affect ABA sensitivity in stomatal guard cells, we attempted to obtain knockout mutants by T-DNA insertion. However, CHLD (At1g08520) is a single gene in Arabidopsis, as is CHLH, and a knockout mutation of CHLD was shown to be albino or lethal (Shen et al. 2006; Huang and Li 2009). Indeed, we could not obtain the homozygous chld mutants (SALK_150219 and SALK_026379). CHLI, however, has two isogenes, designated CHLI1 (At4g18480) and CHLI2 (At5g45930), and CHLI1 is thought to be the major isoform. Consequently, a CHLI1 knockout mutant was able to grow due to the redundant role of CHLI2 (Rissler et al. 2002; Kobayashi et al. 2008; Huang and Li 2009). Indeed, a knockout mutant of CHLI1 (SAIL_230_D11) exhibited a severe dwarf and pale-green phenotype, but we could determine stomatal responses using the isolated epidermis (Fig. 6). In addition, we found that ABA-induced stomatal closure was impaired in this mutant, suggesting that the Mg-chelatase complex as a whole most likely affects the ABA-signaling pathway in guard cells.
Characterization of the T-DNA insertion mutant for Mg-chelatase I subunit 1 (chli1). a Schematic representation of the structure of the CHLI gene and the location of the T-DNA insertion site. Black boxes and lines represent exons and introns, respectively. The T-DNA was inserted into the third exon of the CHLI1 genomic DNA. b CHLI1 expression verification by RT-PCR in the chli1 mutant (upper panel). TUB2 was used as a control (lower panel). c Four-week-old plants, Col and chli1, grown under a 16-h photoperiod. Bar 10 mm. d Effect of ABA on stomatal closing in Col and chli1 plants. Stomatal apertures were measured after incubation for 2.5 h in the basal buffer with (black) or without (white) 20 μM ABA. Other conditions were the same as in Fig. 1d. Data represent the mean with SD. Experiments repeated three times on different occasions gave similar results
Discussion
A screen focused on stomata aperture-dependent water loss via transpiration
One powerful tool for identifying signaling components of stomatal opening and closing is the generation of mutants that show impaired stomatal movements. However, direct microscopic measurement of stomatal apertures is difficult with a large number of plants. Mustilli et al. (2002) reported that the open-stomata mutants, ost1-1 and ost1-2, which were isolated by thermal imaging, exhibit a high rate of water loss via transpiration in their detached leaves. Therefore, measurement of weight changes in detached leaves is an effective method to identify stomatal-aperture mutants. Here, we performed a screen focused on stomatal aperture-dependent water loss in EMS-treated Arabidopsis by weighing detached leaves with a microbalance and successfully isolated three rtl and two stl mutants. To our knowledge, this is the first report of stomatal aperture mutants isolated by this method.
CHLH affects the ABA-signaling pathway in guard cells but is not itself an ABA receptor
Previous reports have suggested that CHLH localized in chloroplasts is an ABA receptor, and that the known missense mutant, cch, has a phenotype that is ABA-insensitive in seed germination, root growth, gene expression, and stomatal movements (Shen et al. 2006; Wu et al. 2009). It should be noted that at the beginning CHLH was identified as an ABA-binding protein using ABA-immobilized at its carboxylate on an affinity resin (Zhang et al. 2002). Given that carboxylate in ABA is needed for bioactivity, this approach potentially possessed a problem (Cutler et al. 2010). In the present study, we could not detect ABA binding to recombinant CHLH protein using 3H-labeled ABA (Fig. 4), and we found no evidence of ABA resistance in either seed germination or root growth in the cch and rtl1 mutants (Fig. S1), even though we performed these experiments in accordance with reported methods (Shen et al. 2006; Melcher et al. 2009; Wu et al. 2009). In contrast, however, the CHLH missense mutants, cch and rtl1 (Figs. 1, 2), as well as CHLH RNAi plants (Fig. 3), showed ABA-insensitive stomatal movements, in agreement with previous reports (Shen et al. 2006; Wu et al. 2009). From these results, we conclude that CHLH specifically affects ABA signaling in guard cells but is not itself an ABA receptor.
The rtl1 mutation is a single nucleotide substitution (C2068 to T), leading to a missense mutation in the protein (L690 to F) (Fig. 2). The missense mutation of cch (P642 to L) is proximally close to that of rtl1, and indeed, both mutants have nearly identical phenotypes. In contrast to these, the missense mutation of gun5-1 (A990 to V), whose phenotype is ABA-sensitive (Shen et al. 2006), is relatively distant from the rtl1 and cch mutations. These results suggest that the region around the rtl1 and cch mutations is important for ABA responsiveness in stomatal guard cells, and that the region around gun5-1 mutation has no effect on the ABA responsiveness. In addition, the loss-of-function mutant, xan-f10, a mutant of Xan-F10 that is an ortholog of CHLH in barley, showed normal ABA responsiveness (Müller and Hansson 2009). However, the mutation of xan-f10 is a 3-bp deletion that removes the conserved amino acid residue E424, suggesting that the region around the xan-f10 mutation has no effect on the ABA responsiveness of stomatal guard cells.
We observed significant stomatal closure in the CHLH missense mutant, rtl1, when ABA was applied simultaneously with a high extracellular concentration of Ca2+ (Fig. 5). Therefore, the CHLH missense mutations of cch and rtl1 may depress Ca2+ mobilization from intracellular Ca2+ stores in response to ABA, thereby damping the cytosolic Ca2+ elevation and/or oscillation in stomatal guard cells. Moreover, these results suggest that chloroplasts may have a crucial role for Ca2+ mobilization since CHLH is a chloroplast-localized protein. It is worthy of note that an important role of chloroplasts for Ca2+ signaling in guard cells has been reported (Nomura et al. 2008; Weinl et al. 2008). Further investigations will be needed to examine intracellular Ca2+ changes in response to ABA in guard cells of rtl1 in the presence and absence of a high concentration of extracellular Ca2+.
Mg-chelatase complex plays an indirect role in ABA signaling in guard cells
Mg-chelatase is a complex enzyme of three subunits, H, D, and I, and all subunits are required for Mg-chelatase activity in chlorophyll biosynthesis (Gibson et al. 1995; Willows et al. 1996; Huang and Li 2009). Our results indicate that not only CHLH missense mutants, but also a CHLI1 knockout mutant, showed ABA insensitivity of stomatal movements (Fig. 6). Therefore, the Mg-chelatase complex as a whole probably affects the ABA-signaling pathway in stomatal guard cells. In addition, Mg-protoporphyrin IX may be involved in the regulation of the ABA signaling in guard cells, since Mg-chelatase complex is Mg-protoporphyrin IX-producing enzyme (Matsuda 2008). Further investigation will be needed to clarify this.
Note that GUN4 is shown to stimulate the activity of Mg-chelatase (Larkin et al. 2003). It is worthy to examine whether GUN4 is involved in ABA signaling in stomatal guard cells, although gun4-1, a missense mutant (Larkin et al. 2003), did not show ABA-insensitive phenotype (Shen et al. 2006).
A possible physiological role of CHLH in the ABA-signaling pathway in guard cells
In the CHLH RNAi experiments, the expression level of CHLH affected the ABA sensitivity of the stomatal guard cells (Fig. 3). This observation is consistent with a report stating that overexpression of a key circadian clock component, TOC1, which interacts with the CHLH promoter and suppresses expression of CHLH, gave rise to a phenotype in which stomatal guard cells were ABA-insensitive (Legnaioli et al. 2009). Taken together, these observations suggest that expression level of CHLH affects the ABA sensitivity of stomatal guard cells. Further investigations of diurnal changes of CHLH expression in guard cells and effects of drought stress on CHLH expression will provide important information on the physiological role of CHLH in the ABA-signaling pathway of stomatal guard cells.
References
Allan AC, Fricker MD, Ward JL, Beale MH, Trewavas AJ (1994) Two transduction pathways mediate rapid effects of abscisic acid in Commelina guard cells. Plant Cell 6:1319–1328
Allen GJ, Kuchitsu K, Chu SP, Murata Y, Schroeder JI (1999) Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid-induced cytoplasmic calcium rises in guard cells. Plant Cell 11:1785–1798
Allen GJ, Chu SP, Schumacher K, Shimazaki CT, Vafeados D, Kemper A, Hawke SD, Tallman G, Tsien RY, Harper JF, Chory J, Schroeder JI (2000) Alteration of stimulus-specific guard cell calcium oscillations and stomatal closing in Arabidopsis det3 mutant. Science 289:2338–2342
Allen GJ, Chu SP, Harrington CL, Schumacher K, Hoffmann T, Tang YY, Grill E, Schroeder JI (2001) A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature 411:1053–1057
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61:651–679
Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KAS, Romeis T, Hedrich R (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase–phosphatase pair. Proc Natl Acad Sci USA 106:21425–21430
Gibson LCD, Willows RD, Kannangara CG, von Wettstein D, Hunter CN (1995) Magnesium-protoporphyrin chelatase of Rhodobacter sphaeroides: reconstitution of activity by combining the products of the bchH, -I and -D genes expressed in Escherichia coli. Proc Natl Acad Sci USA 92:1941–1944
Hayashi Y, Nakamura S, Takemiya A, Takahashi Y, Shimazaki K, Kinoshita T (2010) Biochemical characterization of in vitro phosphorylation and dephosphorylation of the plasma membrane H+-ATPase. Plant Cell Physiol 51:1186–1196
Huang YS, Li H (2009) Arabidopsis CHLI2 can substitute for CHLI1. Plant Physiol 150:636–645
Hwang JU, Lee Y (2001) Abscisic acid-induced actin reorganization in guard cells of dayflower is mediated by cytosolic calcium levels and by protein kinase and protein phosphatase activities. Plant Physiol 125:2120–2128
Inoue S, Kinoshita T, Matsumoto M, Nakayama KI, Doi M, Shimazaki K (2008) Blue light-induced autophosphorylation of phototropin is a primary step for signaling. Proc Natl Acad Sci USA 105:5626–5631
Islam MM, Hossain MA, Jannat R, Munemasa S, Nakamura Y, Mori IC, Murata Y (2010) Cytosolic alkalization and cytosolic calcium oscillation in Arabidopsis guard cells response to ABA and MeJA. Plant Cell Physiol 51:1721–1730
Kim T-H, Böhmer M, Hu H, Nishimura N, Schroeder JI (2010) Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol 61:561–591
Kinoshita T, Shimazaki K (1999) Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J 18:5548–5558
Kobayashi K, Mochizuki N, Yoshimura N, Motohashi K, Hisabori T, Masuda T (2008) Functional analysis of Arabidopsis thaliana isoforms of Mg-chelatase CHLI subunit. Photochem Photobiol Sci 7:1188–1195
Larkin RM, Alonso JM, Ecker JR, Chory J (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299:902–906
Lee SC, Lan W, Buchanan BB, Luan S (2009) A protein kinase–phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA 106:21419–21424
Legnaioli T, Cuevas J, Mas P (2009) TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J 28:3745–3757
Liu X, Yue Y, Li B, Nie Y, Li W, Wu W-H, Ma L (2007) A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science 315:1712–1716
Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324:1064–1068
Marten H, Konrad KR, Dietrich P, Roelfsema MRG, Hedrich R (2007) Ca2+-dependent and -independent abscisic acid activation of plasma membrane anion channels in guard cells of Nicotiana tabacum. Plant Physiol 143:28–37
Matsuda T (2008) Recent overview of the Mg branch of the tetrapyrrole biosynthesis leading to chlorophylls. Photosynth Res 96:121–143
McCourt P, Creelman R (2008) The ABA receptors—we report you decide. Curr Opin Plant Biol 11:474–478
Melcher K, Ng L-M, Zhou X-E, Soon F-F, Xu Y, Suino-Powell KM, Park S-Y, Weiner JJ, Fujii H, Chinnusamy V, Kovach A, Li J, Wang Y, Li J, Peterson FC, Jensen DR, Yong E-L, Volkman BF, Cutler SR, Zhu J-K, Xu HE (2009) A gate-latch-lock mechanism for hormone signaling by abscisic acid receptors. Nature 462:602–608
Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98:2053–2058
Moran R (1982) Formulae for determination of chlorophyllous pigments extracted with N,N-dimethylformamide. Plant Physiol 69:1376–1381
Müller AH, Hansson M (2009) The barley magnesium chelatase 150-kD subunit is not an abscisic acid receptor. Plant Physiol 150:157–166
Mustilli A-C, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14:3089–3099
Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104:34–41
Negi J, Matsuda O, Nagasawa T, Oba Y, Takahashi H, Kawai-Yamada M, Uchimiya H, Hashimoto M, Iba K (2008) CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452:483–486
Nishimura N, Sarkeshik A, Nito K, Park S-Y, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, Yates JR, Schroeder JI (2010) PYR/PYL/RCAR family members are major in vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 61:290–299
Nomura H, Komori T, Kobori M, Nakahira Y, Shiina T (2008) Evidence for chloroplast control of external Ca2+-induced cytosolic Ca2+ transients and stomatal closure. Plant J 53:988–998
Pandey S, Nelson DC, Assmann SM (2009) Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136:136–148
Park S-Y, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow T-F, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu J-K, Schroeder JI, Volkman BF, Cutler SR (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324:1068–1071
Rissler HM, Collakova E, DellaPenna D, Whelan J, Pogson BJ (2002) Chlorophyll biosynthesis. Expression of a second chl I gene of magnesium chelatase in Arabidopsis supports only limited chlorophyll synthesis. Plant Physiol 128:770–779
Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park S-Y, Màrquez JA, Cutler SR, Rodriguez PL (2009) Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 60:575–586
Schroeder JI, Raschke K, Neher E (1987) Voltage dependence of K+ channels in guard-cell protoplasts. Proc Natl Acad Sci USA 84:4108–4112
Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D (2001) Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol 52:627–658
Shang Y, Yan L, Liu Z-Q, Cao Z, Mei C, Xin Q, Wu F-Q, Wang X-F, Du S-Y, Jiang T, Zhang X-F, Zhao R, Sun H-L, Liu R, Yu Y-T, Zhang D-P (2010) The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22:1909–1935
Shen Y-Y, Wang X-F, Wu F-Q, Du S-Y, Cao Z, Shang Y, Wang X-L, Peng C-C, Yu X-C, Zhu S-Y, Fan R-C, Xu Y-H, Zhang D-P (2006) The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443:823–826
Shimazaki K, Doi M, Assmann SM, Kinoshita T (2007) Light regulation of stomatal movement. Annu Rev Plant Biol 58:219–247
Siegel RS, Xue S, Murata Y, Yang Y, Nishimura N, Wang A, Schroeder JI (2009) Calcium elevation-dependent and attenuated resting calcium-dependent abscisic acid induction of stomatal closure and abscisic acid-induced enhancement of calcium sensitivities of S-type anion and inward-rectifying K+ channel in Arabidopsis guard cells. Plant J 59:207–220
Ueno Y, Ishikawa T, Watanabe K, Terakura S, Iwakawa H, Okada K, Machida C, Machida Y (2007) Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell 19:445–457
Vahisalu T, Kollist H, Wang Y-F, Nishimura N, Chan W-Y, Valerio G, Lamminmäki A, Brosche M, Moldau H, Desikan R, Schroeder JI, Kangasjärvi J (2008) SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452:487–491
Weinl S, Held K, Schlücking K, Steinhorst L, Kuhlgert S, Hippler M, Kudla J (2008) A plasmid protein essential for Ca2+-regulated stomatal responses. New Phytol 179:675–686
Willows RD, Gibson LC, Kanangara CG, Hunter CN, von Wettstein D (1996) Three separate proteins constitute the magnesium chelatase of Rhodobacter sphaeroides. Eur J Biochem 235:438–443
Wu F-Q, Xin Q, Cao Z, Liu Z-Q, Du S-Y, Mei C, Zhao C-X, Wang X-F, Shang Y, Jiang T, Zhang X-F, Yan L, Zhao R, Cui Z-N, Liu R, Sun H-L, Yang X-L, Su Z, Zhang D-P (2009) The magnesium-chelatase H subunit binds abscisic acid and functions in abscisic acid signaling: new evidence in Arabidopsis. Plant Physiol 150:1940–1954
Young JJ, Mehta S, Israelsson M, Godoski J, Grill E, Schroeder JI (2006) CO2 signaling in guard cells: calcium sensitivity response modulation, a Ca2+-independent phase, and CO2 insensitivity of the gca2 mutant. Proc Natl Acad Sci USA 103:7506–7511
Zhang D-P, Wu Z-Y, Li X-Y, Zhao Z-X (2002) Purification and identification of a 42-kilodalton abscisic acid-specific-binding protein from epidermis of broad bean leaves. Plant Physiol 128:714–725
Acknowledgments
We thank M. Goto and A. Takaki for their technical support. The cch, gun5-1, and pOCA107-2 were kindly provided by Dr. N. Mochizuki (Kyoto University). This work was supported in part by Grants-in-Aid for Scientific Research (20370021, 22119005, and 21227001) from the Ministry of Education, Culture, Sports, Science and Technology, Japan (T.K.) and by ALCA from the Japan Society for the Promotion of Science (T.K.).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Tsuzuki, T., Takahashi, K., Inoue, Si. et al. Mg-chelatase H subunit affects ABA signaling in stomatal guard cells, but is not an ABA receptor in Arabidopsis thaliana . J Plant Res 124, 527–538 (2011). https://doi.org/10.1007/s10265-011-0426-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-011-0426-x