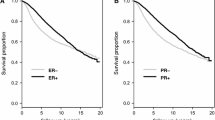
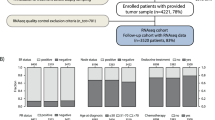
Abstract
To date, three molecular markers (ER, PR, and CYP2D6) have been used in clinical setting to predict the benefit of the anti-estrogen tamoxifen therapy. Our aim was to validate new biomarker candidates predicting response to tamoxifen treatment in breast cancer by evaluating these in a meta-analysis of available transcriptomic datasets with known treatment and follow-up. Biomarker candidates were identified in Pubmed and in the 2007–2012 ASCO and 2011–2012 SABCS abstracts. Breast cancer microarray datasets of endocrine therapy-treated patients were downloaded from GEO and EGA and RNAseq datasets from TCGA. Of the biomarker candidates, only those identified or already validated in a clinical cohort were included. Relapse-free survival (RFS) up to 5 years was used as endpoint in a ROC analysis in the GEO and RNAseq datasets. In the EGA dataset, Kaplan–Meier analysis was performed for overall survival. Statistical significance was set at p < 0.005. The transcriptomic datasets included 665 GEO-based and 1,208 EGA-based patient samples. All together 68 biomarker candidates were identified. Of these, the best performing genes were PGR (AUC = 0.64, p = 2.3E−07), MAPT (AUC = 0.62, p = 7.8E−05), and SLC7A5 (AUC = 0.62, p = 9.2E−05). Further genes significantly correlated to RFS include FOS, TP53, BTG2, HOXB7, DRG1, CXCL10, and TPM4. In the RNAseq dataset, only ERBB2, EDF1, and MAPK1 reached statistical significance. We evaluated tamoxifen-resistance genes in three independent platforms and identified PGR, MAPT, and SLC7A5 as the most promising prognostic biomarkers in tamoxifen treated patients.


Similar content being viewed by others
Abbreviations
- ASCO:
-
American Society of Clinical Oncology
- EGA:
-
European genome–phenome archive
- EGFR:
-
Epidermal growth factor receptor
- ER:
-
Estrogen receptor
- FFPE:
-
Formalin-fixed, paraffin-embedded
- GEO:
-
Gene expression omnibus
- NCCN:
-
National Comprehensive Cancer Network
- NICE:
-
National Institute for Health and Clinical Excellence
- PR:
-
Progesterone receptor
- PRISMA:
-
Preferred reporting items for systematic reviews and meta-analyses
- PROSPERO:
-
International prospective register of systematic reviews
- RFS:
-
Relapse-free survival
- ROC:
-
Receiver operating characteristic
- SABCS:
-
San Antonio breast cancer symposium
- TCGA:
-
The cancer genome atlas
References
Cuzick J, Forbes JF, Sestak I et al (2007) Long-term results of tamoxifen prophylaxis for breast cancer—96-month follow-up of the randomized IBIS-I trial. J Natl Cancer Inst 99:272–282
Davies C, Godwin J, Gray R et al (2011) Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet 378:771–784
Swain SM (2001) Tamoxifen for patients with estrogen receptor-negative breast cancer. J Clin Oncol 19:93S–97S
Early Breast Cancer Trialists’ Collaborative Group (1998) Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet 351:1451–1467
Layfield LJ, Goldstein N, Perkinson KR, Proia AD (2003) Interlaboratory variation in results from immunohistochemical assessment of estrogen receptor status. Breast J 9:257–259
Gyorffy B, Benke Z, Lanczky A et al (2012) RecurrenceOnline: an online analysis tool to determine breast cancer recurrence and hormone receptor status using microarray data. Breast Cancer Res Treat 132:1025–1034
Rakha EA, Reis-Filho JS, Ellis IO (2010) Combinatorial biomarker expression in breast cancer. Breast Cancer Res Treat 120:293–308
Dunnwald LK, Rossing MA, Li CI (2007) Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast Cancer Res 9:R6
Schroth W, Goetz MP, Hamann U et al (2009) Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA 302:1429–1436
Higgins MJ, Stearns V (2011) Pharmacogenetics of endocrine therapy for breast cancer. Annu Rev Med 62:281–293
Visvanathan K, Chlebowski RT, Hurley P et al (2009) American society of clinical oncology clinical practice guideline update on the use of pharmacologic interventions including tamoxifen, raloxifene, and aromatase inhibition for breast cancer risk reduction. J Clin Oncol 27:3235–3258
Moher D, Liberati A, Tetzlaff J, Altman DG (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Open Med 3:e123–e130
Gyorffy B, Schafer R (2009) Meta-analysis of gene expression profiles related to relapse-free survival in 1,079 breast cancer patients. Breast Cancer Res Treat 118:433–441
Gyorffy B, Molnar B, Lage H et al (2009) Evaluation of microarray preprocessing algorithms based on concordance with RT-PCR in clinical samples. PLoS ONE 4:e5645
Li Q, Birkbak NJ, Gyorffy B et al (2011) Jetset: selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics 12:474
Curtis C, Shah SP, Chin SF et al (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486:346–352
Dunning MJ, Smith ML, Ritchie ME, Tavare S (2007) beadarray: R classes and methods for Illumina bead-based data. Bioinformatics 23:2183–2184
Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193
Koboldt DC, Fulton RS, McLellan MD et al (2012) Comprehensive molecular portraits of human breast tumours. Nature 490:61–70
Wickham H (2011) The split-apply-combine strategy for data analysis. J Stat Softw 40:1–29
Gyorffy B, Lanczky A, Eklund AC et al (2010) An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 123:725–731
Gyorffy B, Gyorffy A, Tulassay Z (2005) The problem of multiple testing and solutions for genome-wide studies. Orv Hetil 146:559–563
Musgrove EA, Sutherland RL (2009) Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer 9:631–643
Bartlett JM, Bloom KJ, Piper T et al (2012) Mammostrat as an immunohistochemical multigene assay for prediction of early relapse risk in the tamoxifen versus exemestane adjuvant multicenter trial pathology study. J Clin Oncol 30:4477–4484
Ring BZ, Seitz RS, Beck R et al (2006) Novel prognostic immunohistochemical biomarker panel for estrogen receptor-positive breast cancer. J Clin Oncol 24:3039–3047
Souter S, Lee G (2010) Tubulin-independent tau in Alzheimer’s disease and cancer: implications for disease pathogenesis and treatment. Curr Alzheimer Res 7:697–707
Ikeda H, Taira N, Hara F et al (2010) The estrogen receptor influences microtubule-associated protein tau (MAPT) expression and the selective estrogen receptor inhibitor fulvestrant downregulates MAPT and increases the sensitivity to taxane in breast cancer cells. Breast Cancer Res 12:R43
Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490:61–70
Wu X, Chen H, Parker B et al (2006) HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res 66:9527–9534
Rubin E, Wu X, Zhu T et al (2007) A role for the HOXB7 homeodomain protein in DNA repair. Cancer Res 67:1527–1535
Jin K, Kong X, Shah T et al (2012) The HOXB7 protein renders breast cancer cells resistant to tamoxifen through activation of the EGFR pathway. Proc Natl Acad Sci USA 109:2736–2741
Paruthiyil S, Cvoro A, Tagliaferri M et al (2011) Estrogen receptor beta causes a G2 cell cycle arrest by inhibiting CDK1 activity through the regulation of cyclin B1, GADD45A, and BTG2. Breast Cancer Res Treat 129:777–784
Gee JM, Willsher PC, Kenny FS et al (1999) Endocrine response and resistance in breast cancer: a role for the transcription factor Fos. Int J Cancer 84:54–61
Mendes-Pereira AM, Sims D, Dexter T et al (2012) Genome-wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proc Natl Acad Sci USA 109:2730–2735
Kurebayashi J (2005) Resistance to endocrine therapy in breast cancer. Cancer Chemother Pharmacol 56(Suppl 1):39–46
Gutierrez MC, Detre S, Johnston S et al (2005) Molecular changes in tamoxifen-resistant breast cancer: relationship between estrogen receptor, HER-2, and p38 mitogen-activated protein kinase. J Clin Oncol 23:2469–2476
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Harbeck N, Rody A (2012) Lost in translation? Estrogen receptor status and endocrine responsiveness in breast cancer. J Clin Oncol 30:686–689
Bouzyk M, Gray KP, Regan MM, Pagani O et al (2011) ESR1 and ESR2 polymorphisms in BIG 1-98 comparing adjuvant letrozole (L) versus tamoxifen (T) or their sequence for early breast cancer. J Clin Oncol 29(suppl 27: abstr 1002)
Faraggi D, Simon R (1996) A simulation study of cross-validation for selecting an optimal cutpoint in univariate survival analysis. Stat Med 15:2203–2213
Dignam JJ, Dukic V, Anderson SJ et al (2009) Hazard of recurrence and adjuvant treatment effects over time in lymph node-negative breast cancer. Breast Cancer Res Treat 116:595–602
Saphner T, Tormey DC, Gray R (1996) Annual hazard rates of recurrence for breast cancer after primary therapy. J Clin Oncol 14:2738–2746
Sotiriou C, Wirapati P, Loi S et al (2006) Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 98:262–272
Miller LD, Smeds J, George J et al (2005) An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA 102:13550–13555
Loi S, Haibe-Kains B, Desmedt C et al (2007) Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol 25:1239–1246
Zhang Y, Sieuwerts AM, McGreevy M et al (2009) The 76-gene signature defines high-risk patients that benefit from adjuvant tamoxifen therapy. Breast Cancer Res Treat 116:303–309
Loi S, Haibe-Kains B, Desmedt C et al (2008) Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics 9:239
Desmedt C, Giobbie-Hurder A, Neven P et al (2009) The gene expression grade index: a potential predictor of relapse for endocrine-treated breast cancer patients in the BIG 1–98 trial. BMC Med Genomics 2:40
Symmans WF, Hatzis C, Sotiriou C et al (2010) Genomic index of sensitivity to endocrine therapy for breast cancer. J Clin Oncol 28:4111–4119
Li Y, Zou L, Li Q et al (2010) Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med 16:214–218
Mathieue M (2009) Use of progesterone receptor (PR) expression to predict benefit from prolonged adjuvant tamoxifen (TAM) in breast cancer: Results of a biomarker study from the TAM01 randomized Trial. J Clin Oncol 27(15 suppl: abstr 536)
Klimowicz A (2011) Automated quantification methods improve the accuracy of pr as an independent prognostic factor in tamoxifen treated breast cancer patients. SABCS (abstr P5-11-11)
Tovey S, Dunne B, Witton CJ et al (2005) Can molecular markers predict when to implement treatment with aromatase inhibitors in invasive breast cancer? Clin Cancer Res 11:4835–4842
Zoubir M (2008) Predictive biomarkers for preoperative endocrine therapy of stage II-III breast cancer by tissue microarrays. J Clin Oncol 26(20 suppl: abstr 560)
Bartlett JM, Thomas J, Ross DT et al (2010) Mammostrat as a tool to stratify breast cancer patients at risk of recurrence during endocrine therapy. Breast Cancer Res 12:R47
Hayashida T (2011) Loss of B-cell translocation gene 2 in estrogen receptor–positive breast cancer and tamoxifen resistance. J Clin Oncol 29(suppl 27: abstr 63)
Hilborn E (2011) The importance of CXCL10 and CXCR3-A in breast cancer. SABCS (abstr P1-06-06)
Liu M (2011) Molecular signaling distinguishes early ER positive breast cancer recurrences despite adjuvant tamoxifen SABCS (abstr S1-8)
Palmieri C (2012) Expression of phosphorylated activating transcription factor2 (ATF2) is associated with sensitivity to endocrine therapy in breast cancer SABCS (abstr P3-06-13)
Cannings E, Kirkegaard T, Tovey SM et al (2007) Bad expression predicts outcome in patients treated with tamoxifen. Breast Cancer Res Treat 102:173–179
Millar EK, Anderson LR, McNeil CM et al (2009) BAG-1 predicts patient outcome and tamoxifen responsiveness in ER-positive invasive ductal carcinoma of the breast. Br J Cancer 100:123–133
Lyng M (2012) Gene expression profile that predict outcome of tamoxifen-treated estrogen receptor-positive, high-risk, primary breast cancer patients: a DBCG study. SABCS (abstr P4-09-04)
Larsen M (2011) Bcl-2 as a prognostic marker in breast cancer patients receiving endocrine therapy. SABCS (abstr P2-12-08)
Skvortsov V (2011) Cyclin D1 and its prognostic value in planning of endocrine therapy for women of postmenopausal age with breast cancer. SABCS (abstr P4-01-22)
Surowiak P, Materna V, Paluchowski P et al (2006) CD24 expression is specific for tamoxifen-resistant ductal breast cancer cases. Anticancer Res 26:629–634
Hiscox S (2011) Loss of e-cadherin expression in clinical breast cancer is associated with an adverse outcome on tamoxifen. SABCS (abstr P1-06-18)
Iorns E, Turner NC, Elliott R et al (2008) Identification of CDK10 as an important determinant of resistance to endocrine therapy for breast cancer. Cancer Cell 13:91–104
Shimizu C (2011) Predictive biomarkers of endocrine therapy (ET) for stage IV breast cancer (BC). J Clin Oncol 29(suppl 27: abstr e11090)
Shigekawa T (2012) EBAG9 immunoreactivity is a potential prognostic factor for poor outcome of breast cancer patients with adjuvant tamoxifen therapy SABCS (abstr P6-04-27)
Ejlertsen B, Aldridge J, Nielsen KV et al (2012) Prognostic and predictive role of ESR1 status for postmenopausal patients with endocrine-responsive early breast cancer in the Danish cohort of the BIG 1–98 trial. Ann Oncol 23:1138–1144
Nielsen KV, Ejlertsen B, Muller S et al (2011) Amplification of ESR1 may predict resistance to adjuvant tamoxifen in postmenopausal patients with hormone receptor positive breast cancer. Breast Cancer Res Treat 127:345–355
Singer C (2012) Estrogen receptor alpha (ESR1) gene amplification status and clinical outcome in tamoxifen-treated postmenopausal patients with endocrine-responsive early breast cancer: An analysis of the prospective ABCSG-6 trial. J Clin Oncol 30(suppl: abstr 1050)
Ellsworth R (2012) The effect of HER2 expression on luminal A breast tumors. SABCS (abstr PD02-02)
Reijm EA, Jansen MP, Ruigrok-Ritstier K et al (2011) Decreased expression of EZH2 is associated with upregulation of ER and favorable outcome to tamoxifen in advanced breast cancer. Breast Cancer Res Treat 125:387–394
Reijm E (2012) FOXA1 expression: regulated by EZH2 and associated with favorable outcome to tamoxifen in advanced breast cancer SABCS (abstr P6-04-08)
Hu J (2012) Proteomic screening of FFPE tissue identifies FKBP4 as an independent prognostic factor in hormone receptor positive breast cancers. SABCS (abstr P6-07-17)
Ijichi N, Shigekawa T, Ikeda K et al (2012) Association of double-positive FOXA1 and FOXP1 immunoreactivities with favorable prognosis of tamoxifen-treated breast cancer patients. Horm Cancer 3:147–159
Harada-Shoji N (2012) The role of RIP140 and FOXA1 in breast cancer endocrine sensitivity and resistance. SABCS (abstr P6-04-16)
Reimer T, Koczan D, Muller H et al (2002) Tumour Fas ligand:Fas ratio greater than 1 is an independent marker of relative resistance to tamoxifen therapy in hormone receptor positive breast cancer. Breast Cancer Res 4:R9
Leeb-Lundberg F (2011) G protein-coupled estrogen receptor 1 positively correlates with estrogen receptor a expression and increased distant disease-free survival of breast cancer patients. SABCS (abstr P4-09-02)
Serrero G (2012) Neutralizing antibody to human GP88 (progranulin) restores sensitivity to tamoxifen and inhibits breast tumor growth in mouse xenografts. SABCS (abstr P6-04-19)
De Laurentiis M, Arpino G, Massarelli E et al (2005) A meta-analysis on the interaction between HER-2 expression and response to endocrine treatment in advanced breast cancer. Clin Cancer Res 11:4741–4748
Ma XJ, Wang Z, Ryan PD et al (2004) A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell 5:607–616
Busch S, Ryden L, Stal O et al (2012) Low ERK phosphorylation in cancer-associated fibroblasts is associated with tamoxifen resistance in pre-menopausal breast cancer. PLoS ONE 7:e45669
Watson C, Long JS, Orange C et al (2010) High expression of sphingosine 1-phosphate receptors, S1P1 and S1P3, sphingosine kinase 1, and extracellular signal-regulated kinase-1/2 is associated with development of tamoxifen resistance in estrogen receptor-positive breast cancer patients. Am J Pathol 177:2205–2215
Bergqvist J, Elmberger G, Ohd J et al (2006) Activated ERK1/2 and phosphorylated oestrogen receptor alpha are associated with improved breast cancer survival in women treated with tamoxifen. Eur J Cancer 42:1104–1112
Bianchini G (2011) Molecular tumor characteristics influence adjuvant endocrine treatment outcome. SABCS (abstr S1-7)
Surowiak P, Matkowski R, Materna V et al (2005) Elevated metallothionein (MT) expression in invasive ductal breast cancers predicts tamoxifen resistance. Histol Histopathol 20:1037–1044
Alkner S, Bendahl PO, Grabau D et al (2010) AIB1 is a predictive factor for tamoxifen response in premenopausal women. Ann Oncol 21:238–244
Dihge L, Bendahl PO, Grabau D et al (2008) Epidermal growth factor receptor (EGFR) and the estrogen receptor modulator amplified in breast cancer (AIB1) for predicting clinical outcome after adjuvant tamoxifen in breast cancer. Breast Cancer Res Treat 109:255–262
Zhang L, Gong C, Lau SL et al (2013) SpliceArray profiling of breast cancer reveals a novel variant of NCOR2/SMRT that is associated with tamoxifen resistance and control of ERalpha transcriptional activity. Cancer Res 73:246–255
Bostner J, Ahnstrom Waltersson M, Fornander T et al (2007) Amplification of CCND1 and PAK1 as predictors of recurrence and tamoxifen resistance in postmenopausal breast cancer. Oncogene 26:6997–7005
Bolger J (2012) Global analysis of breast cancer metastasis suggests cellular reprogramming is central to the endocrine resistant phenotype. SABCS (abstr P6-04-01)
Foekens JA, Look MP, Peters HA et al (1995) Urokinase-type plasminogen activator and its inhibitor PAI-1: predictors of poor response to tamoxifen therapy in recurrent breast cancer. J Natl Cancer Inst 87:751–756
Schalper KA (2012) PTEN mRNA positivity using in situ measurements is associated with better outcome in Tamoxifen treated breast cancer patients. SABCS (abstr P3-06-25)
McGlynn LM, Kirkegaard T, Edwards J et al (2009) Ras/Raf-1/MAPK pathway mediates response to tamoxifen but not chemotherapy in breast cancer patients. Clin Cancer Res 15:1487–1495
Basik M (2011) Measurement of Pax2, TC21, CCND1, and RFS1 as predictive biomarkers for outcomes in the NCIC CTG MA.12 trial of tamoxifen after adjuvant chemotherapy in premenopausal women with early breast cancer. J Clin Oncol 29(suppl: abstr 560)
Elsberger B, Paravasthu DM, Tovey SM, Edwards J (2012) Shorter disease-specific survival of ER-positive breast cancer patients with high cytoplasmic Src kinase expression after tamoxifen treatment. J Cancer Res Clin Oncol 138:327–332
Sand-Dejmek J (2011) Wnt5a is a prognostic biomarker in estrogen receptor-positive premenopausal breast cancer. SABCS (abstr P5-01-02)
Acknowledgments
Our work was supported by the OTKA PD 83154 Grant, by the Predict Project (Grant No. 259303 of the EU Health.2010.2.4.1.-8 call), and by the KTIA EU_BONUS_12-1-2013-0003 Grant.
Ethical Standards
We declare that the experiments comply with the current laws of Hungary.
Conflict of interests
There are no disclaimers. The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material. Supplementary R script 1 Supplementary R script 2
Rights and permissions
About this article
Cite this article
Mihály, Z., Kormos, M., Lánczky, A. et al. A meta-analysis of gene expression-based biomarkers predicting outcome after tamoxifen treatment in breast cancer. Breast Cancer Res Treat 140, 219–232 (2013). https://doi.org/10.1007/s10549-013-2622-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-013-2622-y