Abstract
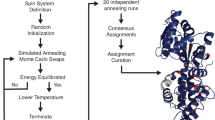
ABACUS [Grishaev et al. (2005) Proteins 61:36–43] is a novel protocol for automated protein structure determination via NMR. ABACUS starts from molecular fragments defined by unassigned J-coupled spin-systems and involves a Monte Carlo stochastic search in assignment space, probabilistic sequence selection, and assembly of fragments into structures that are used to guide the stochastic search. Here, we report further development of the two main algorithms that increase the flexibility and robustness of the method. Performance of the BACUS [Grishaev and Llinás (2004) J Biomol NMR 28:1–101] algorithm was significantly improved through use of sequential connectivities available from through-bond correlated 3D-NMR experiments, and a new set of likelihood probabilities derived from a database of 56 ultra high resolution X-ray structures. A Multicanonical Monte Carlo procedure, Fragment Monte Carlo (FMC), was developed for sequence-specific assignment of spin-systems. It relies on an enhanced assignment sampling and provides the uncertainty of assignments in a quantitative manner. The efficiency of the protocol was validated on data from four proteins of between 68–116 residues, yielding 100% accuracy in sequence specific assignment of backbone and side chain resonances.







Similar content being viewed by others
References
Bailey-Kellogg C, Widge A, Kelley JJ, Berardi MJ, Bushweller JH, Donald BR (2000) The NOESY jigsaw: automated protein secondary structure and main-chain assignment from sparse, unassigned NMR data. J Comput Biol 7:537–558
Bartels C, Xia T, Billeter M, Güntert P, Wüthrich K (1995) The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR 6:1–10
Berg BA, Neuhaus T (1991) Multicanonical algorithms for first order phase transitions. Phys Lett B 267:249–253
Berg BA, Celik T (1992) New approach to spin-glass simulation. Phys Rev Lett 69:2292–2295
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Ferrenberg AM, Swendsen RH (1989) New Monte Carlo technique for studing phase transitions. Phys Rev Lett 63:1658
Goddard TD, Kneller DG (2003) Sparky - NMR assignment and integration software. University of California, San Francisco
Grishaev A, Llinás M (2002a) CLOUDS, a protocol for deriving a molecular proton density via NMR. Proc Natl Acad Sci USA 99:6707–6712
Grishaev A, Llinás M (2002b) Protein structure elucidation from NMR proton density. Proc Natl Acad Sci USA 99:6713–6718
Grishaev A, Llinás M (2004) BACUS: a Bayesian protocol for the identification of protein NOESY spectra via unassigned spin systems. J Biomol NMR 28:1–10
Grishaev A, Steren CA, Wu B, Pineda-Lucena A, Arrowsmith CH, Llinás M (2005) ABACUS, a direct method for protein NMR structure computation via assembly of fragments. Proteins: Struct Funct Genet 61:36–43
Güntert P, Mumenthaler C, Wüthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273:283–298
Güntert P (2004) Automated NMR structure calculation with CYANA. Methods Mol Biol 278:353–378
Hansmann UH, Okamoto Y (1993) Prediction of peptide conformation by multicanonical algorithm: new approach to the multiple-minima problem. J Comp Chem 14:1333–1338
Herrmann T, Güntert P, Wüthrich K (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319:209–227
Kaustov L, Lukin J, Lemak A, Duan S, Ho M, Doherty R, Penn LZ, Arrowsmith CH (2007) The conserved CPH domains of Cul7 and PARC are protein-protein interaction modules that bind the tetramerization domain of p53. J Biol Chem 282:11300–11307
Malmodin D, Billeter M (2005) High-throughput analysis of protein NMR spectra. Progr NMR Spectrosc 46:109–129
Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E (1953) Equation of state calculations by fast computing machines. J Chem Phys 21:1087–1092
Moseley HN, Montelione GT (1999) Automated analysis of NMR assignments and structures for proteins. Curr Opin Struct Biol 9:635–642
Srisailam S, Lukin JA, Lemak A, Yee A, Arrowsmith CH (2006) Sequence Specific resonance assignment of a hypothetical protein PA0128 from pseudomonas aeruginosa. J Biomol NMR 36:27
Wand AJ, Nelson SJ (1991) Refinement of the main chain directed assignment strategy for the analysis of 1H NMR spectra of proteins. Biophys J 59:1101–1112
Wang X, Srisailam S, Yee A, Lemak A, Arrowsmith CH, Prestergard JH, Tian F (2007) Domain-domain motions in proteins from time-modulated pseudocontact shifts. J Biomol NMR 39:53–61
Word JM, Lovell SC, Richardson JS, Richardson DC (1999) Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J Mol Biol 285:1735–1747
Wu B, Yee A, Pineda-Lucena A, Semesi A, Ramelot TA, Cort JR, Jung JW, Edwards A, Lee W, Kennedy M, Arrowsmith CH (2003) Solution structure of ribosomal protein S28E from methanobacterium thermoautotrophicum. Protein Sci 12:2831–2837
Yee A, Chang X, Pineda-Lucena A, Wu B, Semesi A, Le B, Ramelot T, Lee GM, Bhattacharyya S, Gutierrez A, Denisov A, Lee C, Cort JR, Kozlov G, Liao J, Finak G, Chen L, Wishart D, Lee W, McIntosh LP, Gehring K, Kennedy MA, Edwards AM, Arrowsmith CH (2002) An NMR approach to structural proteomics. Proc Nat Acad Sci USA 99:1825–1830
Zimmerman DE, Montelione GT (1995) Automated analysis of nuclear magnetic resonance assignment for proteins. Curr Opin Struct Biol 5:664–673
Acknowledgements
We wish to thank Dr. Lilia Kaustov and Dr. Sampas Srisailam for providing us with resonance assignments of CPH domain of Cul7 and protein pa0128, respectively, which were performed by conventional manual procedure. We also thank Dr. Bin Wu for providing us with complete resonance assignment and NOESY peak lists of proteins mth1743 and mth0256. This work was supported by the Ontario Research and Development Challenge Fund, the US National Institute of Health Protein Structure Initiative (P50-GM62413-01 and GM67965) through the Northeast Structural Genomics Consortium, Genome Canada through the Ontario Genomics Institute, and The Canadian Institutes of Health Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lemak, A., Steren, C.A., Arrowsmith, C.H. et al. Sequence specific resonance assignment via Multicanonical Monte Carlo search using an ABACUS approach. J Biomol NMR 41, 29–41 (2008). https://doi.org/10.1007/s10858-008-9238-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-008-9238-2