Abstract
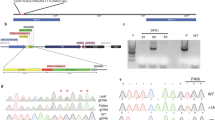
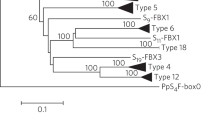
Tetraploid sour cherry (Prunus cerasus) exhibits a genotype-dependent loss of gametophytic self-incompatibility that is caused by the accumulation of non-functional S-haplotypes with disrupted pistil component (stylar-S) and/or pollen component (pollen-S) function. Genetic studies using diverse sour cherry germplasm identified non-functional S-haplotypes for which an equivalent wild-type S-haplotype was present in sweet cherry (Prunus avium), a diploid progenitor of sour cherry. In all cases, the non-functional S-haplotype resulted from mutations affecting the stylar component S-RNase or Prunus pollen component S-haplotype-specific F-box protein (SFB). This study determines the molecular bases of three of these S-haplotypes that confer unilateral incompatibility, two stylar-part mutants (S 6m2 and S 13m ) and one pollen-part mutant (S 13 ′). Compared to their wild-type alleles, S 6m2 -RNase has a 1 bp deletion, S 13m -RNase has a 23 bp deletion and SFB 13 ′ has a 1 bp substitution that lead to premature stop codons. Transcripts were identified for these three alleles, S 6m2 -RNase, S 13m -RNase, and SFB 13 ′, however, these transcripts presumably result in altered proteins with a resulting loss of activity. Our characterization of natural pollen-part and stylar-part mutants in sour cherry along with other natural S-haplotype mutants identified in Prunus supports the view that loss of pollen specificity and stylar rejection evolve independently and are caused by structural alterations affecting the S-haplotype. The prevalence of non-functional S-haplotypes in sour cherry but not in sweet cherry (a diploid) suggests that polyploidization and gene duplication were indirectly responsible for the dysfunction of some S-haplotypes and the emergence of self-compatibility in sour cherry. This resembles the specific mode of evolution in yeast where accelerated evolution occurred to one member of the duplicated gene pair.
Similar content being viewed by others
Abbreviations
- GSI:
-
Gametophytic self-incompatibility
- RT-PCR:
-
Reverse transcriptase PCR
- R:
-
Rheinische
- SI:
-
Self-incompatible
- SC:
-
Self-compatible
- SFB:
-
S-haplotype-specific F-box protein
References
Anderson MA, Cornish EC, Mau S-L, Williams EG, Hoggart R, Atkinson A, Bonig I, Grego B, Simpson R, Roche P, Haley JD, Penschow J, Niall HD, Tregear GW, Coghlan JP, Crawford RJ, Clarke AE (1986) Cloning of cDNA for a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata. Nature 321:38–44
Beaver JA, Iezzoni AF (1993) Allozyme inheritance in tetraploid sour cherry (Prunus cerasus L.). J Am Soc Hort Sci 111:873–877
Bošković RI, Wolfram B, Tobutt KR, Cerović R, Sonneveld T (2006) Inheritance and interactions of incompatibility alleles in the tetraploid sour cherry. Theor Appl Genet 112:315–326
Brettin TS, Karle R, Crowe EL, Iezzoni AF (2000) Chloroplast inheritance and DNA variation in sweet, sour, and ground cherry. J Hered 91:75–79
Brewbaker JL, Natarajan AT (1960) Centric fragments and pollen part mutation of incompatibility alleles in Petunia. Genetics 45:699–704
de Nettancourt D (2001) Incompatibility and incongruity in wild and cultivated plants. Springer, Berlin
Entani T, Iwano M, Shiba H, Che F-S, Isogai A, Takayama S (2003) Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen- expressed F-box gene with allelic diversity. Genes Cells 8:203–213
Golz JF, Oh H-Y, Su V, Kusaba M, Newbigin E (2001) Genetic analysis of Nicotiana pollen-part mutants is consistent with the presence of an S-ribonuclease inhibitor at the S locus. Proc Natl Acad Sci USA 98:15372–15376
Golz JF, Su V, Clarke AE, Newbigin E (1999) A molecular description of mutations affecting the pollen components of the Nicotiana alata S locus. Genetics 152:1123–1135
Hauck NR, Yamane H, Tao R, Iezzoni AF (2002) Self-compatibility and incompatibility in tetraploid sour cherry (Prunus cerasus L.). Sex Plant Reprod 15:39–46
Hauck NR, Yamane H, Tao R, Iezzoni AF (2006) Accumulation of non-functional S-haplotypes results in the breakdown of gametophytic self-incompatibility in tetraploid Prunus. Genetics 172:1191–1198
Iezzoni A, Schmidt H, Albertini A (1990) Cherries (Prunus spp.). In: Moore JN (eds) Genetic resources of temperate fruit and nut crops. ISHS, Wageningen, Netherlands, pp 110–173
Ikeda K, Igic B, Ushijima K, Yamane H, Hauck NR, Nakano R, Sassa H, Iezzoni AF, Kohn JR, Tao R (2004) Primary structure futures of the S haplotype-specific F-box protein, SFB, in Prunus. Sex Plant Reprod 16:235–243
Ikeda K, Ushijima K, Yamane H, Tao R, Hauck NR, Sebolt AM, Iezzoni AF (2005) Linkage and physical distances between the S-haplotype S-RNase and SFB genes in sweet cherry. Sex Plant Reprod 17:289–296
Kao T-H, Tsukamoto T (2004) The molecular and genetic basis of S-RNase-based self-incompatibility. Plant Cell 16(Suppl):S72–S83
Kellis M, Birren BW, Lander ES (2004) Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428:617–624
Lansari A, Iezzoni A (1990) A preliminary analysis of self-incompatibility in sour cherry. Hort Sci 25:1636–1638
Lewis D (1949) Structure of the self-incompatibility gene. II. Heredity 3:339–355
Matthews P, Lapins K (1967) Self-fertile sweet cherries. Fruit Var Hort Digest 21:36–37
McClure B, Mou B, Canevascini S, Bernatzky R (1999) A small asparagine-rich protein required for S-allele-specific pollen rejection in Nicotiana. Proc Natl Acad Sci 96:13548–13553
Olden EJ, Nybom N (1968) On the origin of Prunus cerasus L. Hereditas 59:327–345
Pandy KK (1965) Centric chromosome fragments and pollen-part mutation of the incompatibility gene in Nicotiana alata. Nature 206:792–795
Qiao H, Wang F, Zhao L, Zhou J, Lai Z, Zhang Y, Robbins TP, Xue Y (2004) The F-box protein AhLF-S2 controls the pollen function of S-RNase-based self-incompatibility. Plant Cell 16:2307–2322
Royo J, Kuntz C, Kowyama Y, Anderson MA, Clarke AE, Newbigin E (1994) Loss of a histidine residue at the active site of the S-locus ribonuclease is associated with self-compatibility in Lycopersicon peruvianum. Proc Natl Acad Sci USA 91:6511–6514
Sassa H, Hirano H, Ikehashi H (1992) Self-incompatibility-related RNases in the style of Japanese pear (Pyrus serotina Rehd.). Plant Cell Physiol 33:811–814
Sassa H, Nishio T, Kowyama Y, Hirano H, Koba T, Ikehashi H (1996) Self-incompatibility (S) alleles of the Rosaceae encode members of a distinct class of the T2/S ribonuclease superfamily. Mol Gen Genet 250:547–557
Sijacic P, Wang X, Skirpan AL, Wang Y, Dowd PE, McCubbin AG, Huang S, Kao T-H (2004) Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429:302–305
Sonneveld T, Robbins TP, Bošković R, Tobutt KR (2001) Cloning of six cherry self-incompatibility alleles and development of allele-specific PCR detection. Theor Appl Genet 102:1046–1055
Sonneveld T, Tobutt KR, Vaughan SP, Robbins TP (2005) Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. Plant Cell 17:37–51
Tao R, Yamane H, Sugiura A, Murayama H, Sassa H, Mori H (1999) Molecular typing of S-alleles through identification, characterization and cDNA cloning for S-RNases in sweet cherry. J Am Soc Hort Sci 124:224–233
Tobutt KR, Bošković R, Cerović R, Sonneveld T, Ruzić D (2004) Identification of incompatibility alleles in the tetraploid species sour cherry. Theor Appl Genet 108:775–785
Tsukamoto T, Ando T, Watanabe H, Marchesi E, Kao T-H (2005) Duplication of the S-locus F-box gene is associated with breakdown of pollen function in an S-haplotype identified in a natural population of self-incompatible Petunia axillaris. Plant Mol Biol 57:141–153
Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H (2003) Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15:771–781
Ushijima K, Sassa H, Kusaba M, Tao R, Tamura M, Gradziel TM, Dandekar AM, Hirano H (2001) Characterization of the S-locus region of almond (Prunus dulcis): analysis of a somaclonal mutant and a cosmid contig for an S haplotype. Genetics 158:379–386
Ushijima K, Sassa H, Tao R, Yamane H, Dandekar AM, Gradziel TM, Hirano H (1998) Cloning and characterization of cDNAs encoding S-RNases in almond (Prunus dulcis): primary structure features and sequence diversity of the S-RNases in Rosaceae. Mol Gen Genet 260:261–268
Ushijima K, Yamane H, Watari A, Kakehi E, Ikeda K, Hauck NR, Iezzoni AF, Tao R (2004) The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J 39:573–586
Vaughan SP, Russell K, Sargent DJ, Tobutt KR (2006) Isolation of S-locus F-box alleles in Prunus avium and their application in a novel method to determine self-incompatible genotype. Theor Appl Genet 112:856–866
Voytas DF, Naylor GJP (1998) Rapid flux in plant genomes. Nat Genet 20:6–7
Wünsch A, Hormaza JI (2004a) Cloning and characterization of genomic DNA sequences of four self-incompatibility alleles in sweet cherry (Prunus avium L.). Theor Appl Genet 108:299–305
Wünsch A, Hormaza JI (2004b) Genetic and molecular analysis in Cristobalina sweet cherry, a spontaneous self-compatible mutant. Sex Plant Reprod 17:203–210
Xue Y, Carpenter R, Dickinson HG, Coen ES (1996) Origin of allelic diversity in Antirrhinum S locus RNases. Plant Cell 8:805–814
Yamane H, Tao R, Sugiura A, Hauck NR, Iezzoni AF (2001) Identification and characterization of S-RNases in tetraploid sour cherry (Prunus cerasus). J Am Soc Hort Sci 126:661–667
Yamane H, Ikeda K, Hauck NR, Iezzoni AF, Tao R (2003a) Self-incompatibility (S) locus region of the mutated S 6-haplotypes of sour cherry (Prunus cerasus) contains a functional pollen S-allele and a non-functional pistil S allele. J Exp Bot 54:2431–2434
Yamane H, Ikeda K, Ushijima K, Sassa H, Tao R (2003b) A pollen-expressed gene for a novel protein with an F-box motif that is very tightly linked to a gene for S-RNase in two species off cherry, Prunus cerasus and P. avium. Plant Cell Physiol 44:764–769
Yamane H, Ushijima K, Sassa H, Tao R (2003c) The use of the S haplotype-specific F-box protein gene, SFB, as a molecular marker for S-haplotypes and self-compatibility in Japanese apricot (Prunus mume). Theor Appl Genet 107:1357–1361
Zhao X, Si Y, Hanson RE, Crane CF, Price HJ, Stelly DM, Wendel JF, Paterson AH (1998) Dispersed repetitive DNA has spread to new genomes since polyploidy formation in cotton. Genome Res 8:479–492
Acknowledgments
This work was supported by a grant from the USDA Cooperative State Research, Education, and Extension Service—National Research Initiative—Plant Genome Program.
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequences reported in this paper have been submitted to the GenBank/EMBL/DDBJ database under accession numbers: DQ385841 (S 6m2 -RNase), DQ385842 (S 13 -RNase), DQ385843 (S 13m -RNase), DQ385844 (SFB 13 ), and␣DQ385845 (SFB 13 ′)
Rights and permissions
About this article
Cite this article
Tsukamoto, T., Hauck, N.R., Tao, R. et al. Molecular characterization of three non-functional S-haplotypes in sour cherry (Prunus cerasus). Plant Mol Biol 62, 371–383 (2006). https://doi.org/10.1007/s11103-006-9026-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-006-9026-x