Abstract
The CCN family of proteins (CCN1, CCN2, CCN3, CCN4, CCN5 and CCN6) are multifunctional mosaic proteins that play keys roles in crucial areas of physiology such as angiogenesis, skeletal development tumourigenesis, cell proliferation, adhesion and survival. This expansive repertoire of functions comes through a modular structure of 4 discrete domains that act both independently and in concert. How these interactions with ligands and with neighbouring domains lead to the biological effects is still to be explored but the molecular structure of the domains is likely to play an important role in this. In this review we have highlighted some of the key features of the individual domains of CCN family of proteins based on their biological effects using a homology modelling approach.
Similar content being viewed by others
Introduction
The CCN family of proteins is a crucial group of signalling molecules found in eukaryotic organisms. The CCN nomenclature is due to the first three and most well studied members of the family: Cyr61 (Cysteine rich protein 61), CTGF (Connective tissue growth factor) and NOV (Nephroblastoma overexpressed gene) (Bork 1993) which are now designated CCN1, CCN2 and CCN3 (Brigstock et al. 2003). Three other family members (CCN4/WISP1, CCN5/WISP2 and CCN6/WISP3) have been described. Each of the CCN molecules was given several other names relating to other biological activities that they possess based on the wide range of biological functions they are involved in including- adhesion, mitogenesis, migration and chemotaxis, cell survival, differentiation, angiogenesis, chondrogenesis, tumourigenesis and wound healing making them an important group of molecules to study (Brigstock et al. 2003).
The full scope of the CCN proteins’ biological function is beyond the scope of this article but details can be found in a wide range of reviews (Lau and Lam 1999; Brigstock 1999; Perbal 2001a; Perbal 2004; Perbal and Takigawa 2005; Leask and Abraham 2006).
The multi-domain structure of the CCN proteins
Like many extracellular matrix (ECM) proteins the CCN family of proteins (Table 1) is constructed from a series of discrete domains built from the library of known eukaryotic domains (Hohenester and Engel 2002). The domains in the CCN molecules have been identified through sequence alignment or conserved amino acid motifs (Bork 1993) although the exact biological role of each module is still not fully understood. We have constructed structural models of each of the individual domains and tried to correlate them with the known functional aspects of each domain.
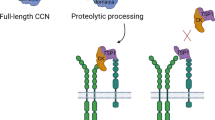
The CCN proteins all share the same modular structure of 4 well ordered and discrete domains that are stable as small truncates or individually (Kubota et al. 2006) and are closely related on the amino acid level with ~30–50% identity (40–60% similarity) (Brigstock et al. 2003; Perbal and Planque 2006; Kubota and Takigawa 2006; Schuetze et al. 2006). The protein is threaded in 4 domains at both protein and DNA levels with an exon corresponding to the signal sequence followed by 4 more exons, one per domain. Many large multi-modular human proteins are built in this way from blocks of exons and it is thought that many mosaic extracellular proteins have come about through evolutionary shuffling of these basic building blocks (Bork 1993; Bornstein 1995; Kireeva et al. 1996). A sequence alignment of the CCN proteins using the T-Coffee program (Notredame et al. 2000) highlighting the conserved cysteine residues and other notable parts of each domain is shown below in Fig. 1 (Holbourn et al. 2008). In the CCN proteins the domain following an N-terminal secretory signal peptide are: (i) An insulin-like growth factor binding protein —like domain (IGFBP), (ii) A von Willebrand factor type C repeat module (VWC), (iii) A Thrombospondin type-1 repeat module (TSP1) and (iv) A Cysteine knot containing module (CT). A representation of each domain and their known binding partners are shown in Fig. 2 (Bork 1993). CCN5 is the exception to this arrangement as it lacks the 4th CT domain. Another notable feature of the CCN proteins is their high cysteine content, almost 10% by mass that includes 38 cysteines divided into 17 potential conserved disulphide bonds spread across the 4 domains, except CCN5 that lacks 10 cysteines in its missing CT domain and CCN6 that lacks 4 cysteines in the VWC domain. The entire molecule can also be split into two halves, an N-terminal half with the IGFBP and VWC domains and the C-terminal half with the TSP and CT domains, separated by a flexible and protease vulnerable linker (Bork 1993; Lau and Lam 1999). This linker region while varying greatly in length and amino acid composition does have several “L/I/Y R/V” sites that are vulnerable to proteolysis by a selection of matrix metalloproteases (MMPs) (Hashimoto et al. 2002). It has been shown that a wide variety of matrix metalloproteases (MMP-1, 2, 3, 7, 9 ,13) target this central linker region and additional proteases such as elastase and plasmin could attack linkers that connect domains 1 and 2 or domains 3 and 4 (de Winter et al. 2008; Hashimoto et al. 2002; Brigstock et al. 1997; Ball et al. 1998). Cleavage at these sites can give bioactive truncated components that can be used as markers in some types of disease including some fibrotic diseases (N-terminal fragments) (Gao and Brigstock 2004) and some types of pancreatic cancer (C-terminal fragments) (Gao and Brigstock 2006). These truncated components can show distinct biological properties and might constitute an additional process for the regulation of the biological activity of the CCN proteins and in some cases to be associated with some of their functions (Perbal 2001b; Perbal 2004; Tong and Brigstock 2006; Brigstock 1999).
Sequence alignment of the CCN proteins. The sequences of the 6 human CCN proteins were aligned using the T-Coffee server (Notredame et al 2000). The start and finish points of each domain are indicated and various important regions on the molecules are highlighted. In the IGFBP domain the “thumb” and GCGCCxxC motif are shaded in red. In the VWC domain the integrin αvβ3 site is highlighted in yellow. In the TSP domain the α6β1 binding site is highlighted in blue. This region also contains the bioactive peptide section (Karagiannis and Popel 2007). The integrin α6β1 and HSPG binding sites and the possible reverse integrin GDR motif in the CT domain are all highlighted in green
Details of the individual domains . Close-up views of each of the domains using the model domains of CCN1. The known substrates for each domain are listed including: insulin like growth factors (IGFs); bone morphogenic proteins (BMPs); transforming growth factor –β (TGF); LDL receptor protein 1 (LRP1) and heparin sulphated proteoglycans (HSPGs). In the IGFBP domain the two subdomains can be seen and the ladder of disulphide bonds in the “palm” sub-domain. In the VWC domain the two sub-domains can be seen; the more structured N-terminal domain and the less structured C-terminal domain in a fibronectin like fold. In the TSP domain the 3 strands are visible and the CWR layers are labelled. In the partial model of the CT domain the cysteine knot can be seen although the heparin binding N-terminal tips cannot be seen. An unbound cysteine protrudes through the centre of each knot though it is unknown if this is used to form dimers or a disulphide bridge with another cysteine in the missing (not modelled) section of the CT domain
Whilst each module no doubt has its own specific biological role many of the functional effects of the CCN family come about through multiple modules acting in concert. Truncated proteins, or proteins missing internal modules, have been shown to possess different biological activities, and in some cases be associated to pathological situations (Perbal 2001b). An example of the multi-domain requirements of some functions has been seen in CCN2 where individual modules could replicate some of the effects of full length CCN2 but for other functions, such as p38 MAPK activation the full length protein or a cocktail containing the 4 individual domains was required (Kubota et al. 2006). The ability of the 4 individual modules to give rise to the same biological behaviour as the full length protein suggest that there must be some sort of cumulative physical interaction between multiple modules and the substrates.
The IGFBP domain
There are 6 members in the human insulin-like growth factor binding protein (IGFBP) family and they are involved in several important biological functions that are centred around modulating insulin-like growth factors (IGFs): 1) to act as a transport proteins for the IGFs, 2) to enable localisation of IGF availability, 3) to regulate the metabolic break down of IGFs and extend their biological lifespan and 4) to directly affect the interaction between IGFs and their receptors on the cell surface and in doing so indirectly control IGF function (Jones and Clemmons 1995). The indirect control of IGF function extends the role of the IGFBP proteins into many diverse areas of cellular functions including: amino acid and glucose uptake, cell cycle progression, cell proliferation; cell death, cell differentiation; chemotaxis, hormone and neurotransmitter secretion, and parts of the immune response (Jones and Clemmons 1995). Recently modulation of IGF function was shown to play a critical role in aggressive inflammatory breast cancers and in a wide variety of other cancers (Helle 2004). The strong link between IGF regulation and cancer could be important for CCN4-6 that are heavily involved in aggressive inflammatory breast cancer and exert their influence via control and manipulation of IGF-1 (Zhang et al. 2005; Davies et al 2007). Whether this interaction is as a result of the IGFBP domains of CCN4-6 interacting directly with IGF or indirectly through other pathways is unknown.
The IGFBP have several distinguishing features in addition to a strong sequence similarity and conserved motifs. They are cysteine rich bi-domain proteins with their N- and C-terminal domains linked by a short flexible region. The linker region varies between IGFBPs and similar to the inter-domain linkers in the CCN family and has several sites vulnerable to protease degradation (Hwa et al. 1999; Firth and Baxter 2002). There are a total of 18 cysteines, 12 in the N-terminal domain and 6 in the C-terminal domain that are all potentially involved in internal disulphide bond formation within each domain. The N-terminal domain is the domain that is found in the CCN family and is a globular domain that contains one of the conserved motifs as well as the IGF binding cavity (Kalus et al. 1998; Hwa et al. 1999). Although the N-terminal domain contains the IGF binding cavity it is the jaw-like structure of the N- and C-termini surrounding the IGF molecule that leads to high affinity (KD ~1nM) binding. Importantly, it has been reported that the N and C terminal regions can bind IGF independently in some of the IGFBPs, as would be the case for the IGFBP domain in the CCN proteins (Kim et al. 1997) though when working independently the IGF binding is several orders of magnitude lower than the full length protein (Stndker et al. 2000).
The CCN proteins have been classified by some as additional IGFBPs or as IGFBP-related proteins (IGFBP-rPs) (Hwa et al. 1999; Kim et al. 1997) due to their high degree of sequence homology to the N-terminal region of the traditional IGFBPs. Though the CCN domains’ IGF affinity is hundred fold lower than the full length IGFBPs it is in the same range as the binding seen for N-terminal truncates of the IGFBPs (Kim et al 1997; Yamanaka et al. 1997). The reduced IGF affinity, due to the lack of a C-terminal domain in CCN proteins has led to two classes of IGFPBs; high and low affinity IGFBPs. In this classification, the 6 traditional IGFBPs are considered high affinity and the CCN proteins (IGFBP-rPs) and other proteins that only contain an N-terminal IGFPB domain are considered low affinity binders (Hwa et al 1999; Kim et al 1997).
In the CCN family there is little information on the exact role played by the IGFBP domain in CCN function. The binding of CCN domains to IGF is still not fully characterised as in some experiments IGF binding by CCN IGFBP domain has been found to be lacking and chimeras with the C-terminal domain of IGFBP3 fused to the IGFBP domain of CCN3 have shown lack of binding (Yan et al. 2006). Although its binding and interactions with IGF are for the most part unknown and subject to considerable debate, it has been shown that the independent IGFBP domain is biologically active in other cellular pathways (Kubota et al. 2006). The IGFBP domain of CCN2 is capable of stimulating JNK mediated proliferation, in contrast to the other domains promoting differentiation, and it is the only independent domain that was unable to promote ERK signalling (Kubota et al. 2006). In healthy cells the IGFBP domain of CCN6 is thought to be involved in regulating IGF-1 availability (Perbal 2003; Zhang et al. 2005). This IGF regulation may prove to be important in cancer as IGF is a key regulatory molecule and its deregulation can lead to several types of severe cancer (Zhang et al. 2005; Davies et al. 2007). CCN6 has been seen to be knocked out in 80% of cases of aggressive inflammatory breast cancer and results in uncontrolled IGF1 induced cell growth and tumourigenesis. Functional CCN6 has been seen to limit the invasive and motile effects of unregulated IGF that can lead to aggressive inflammatory breast cancer (Zhang et al. 2005) though the exact mode through which it inhibits tumour growth is unknown. This role in breast cancer mediated through IGF function is shared to a lesser extent with CCN4 and 5 (Zhang et al. 2005). In addition CCN4–6 can now be used as prognostic markers for breast and other cancer leading suggesting the IGFBP domain plays an important, and as yet not fully understood role in tumourigenesis (Davies et al. 2007).
The CPH model server (Lund et al. 2002) was used to construct models of the IGFBP domains of CCN1, 2, 3 and 6, which are shown in Figs. 3, 4, 5 and 6, using the 79 amino acid NMR structure of IGFBP4 as the template with which they share ~30% sequence identity [PDB code 1dsp] (Sitar et al. 2006) and a high level of similarity. The structure of IGFBP4 N-terminal has a roughly L-shaped appearance and can be divided into two perpendicular subdomains connected by a short stretch of coil and has been compared to a hand with a “palm”, “thumb” and “finger” sections. The first of these subdomains consists of a 2-stranded β-sheet adjacent to a ladder of parallel loops of coil stabilised by 3 disulphide bonds that are in the same plane forming the flat palm of the molecule and can be seen in Fig. 2. This is a structural segment of the domain as it is not involved in IGF binding (Kalus et al 1998; Sitar et al. 2006). In some of the traditional IGFBPs such as IGFBP4 (Sitar et al. 2006) the very N-terminal residues protrude forming a “thumb” on the IGFBP binding domain that can play a role in binding to the IGFs by wrapping around the IGF molecule and forming hydrophobic interactions with aromatic residues of the IGF molecule to increase the affinity (Sitar et al. 2006). However these thumb segments are not seen in sequence alignments with the CCN IGFBP domains. The second subdomain, forms the “fingers” of the N-terminal domain, is a globular domain centred around a 3 stranded anti-parallel β-sheet strengthened by an internal disulphide bridge that links strands 1 and 3. The IGF binding site is formed in this small subdomain forming a cleft lined by mainly hydrophobic residues that comfortably accommodate a large hydrophobic patch on the IGF molecule (Kalus et al. 1998; Siwanowicz et al. 2005; Sitar et al. 2006). The strongly conserved GCGCCxxC motif is not actually involved directly in IGF binding instead being a structural motif that enables the globular domain to form a rigid base that supports and separates the thumb sequence and “fingers” to keep them in correct positions to bind the IGF molecule (Sitar et al 2006).
Model of CCN1. The model of the 4 domains of CCN1 is shown in ribbon and surface representations arranged in N- to C- termini from top to bottom. The linker regions with their variable sequence and flexible nature could not be modelled and it is unknown if there are intra-domain interactions about a hinge. This figure was adapted from Holbourn et al (2008) with permission from the authors
Model of CCN2. The model of the 4 domains of CCN2 is shown in ribbon and surface representations arranged in N- to C- termini from top to bottom. The linker regions with their variable sequence and flexible nature could not be modelled and it is unknown if there are intra-domain interactions about a hinge. Unlike the others the CT domain of CCN2 has a predicted short α-helix in place of a β-strand. This figure was adapted from Holbourn et al (2008) with permission from the authors
Model of CCN3. The model of the 4 domains of CCN3 is shown in ribbon and surface representations arranged in N- to C- termini from top to bottom. The linker regions with their variable sequence and flexible nature could not be modelled and it is unknown if there are intra-domain interactions about a hinge. This figure was adapted from Holbourn et al (2008) with permission from the authors
Models of CCN 4–6. The partial models of CCN4–6 are shown in ribbon and surface representations arranged in N- to C- termini from top to bottom. The linker regions with their variable sequence and flexible nature could not be modelled and it is unknown if there are intra-domain interactions about a hinge. The question marks represent domains that were unable to be modelled. The IGFBP domain of CCN6 shows considerable differences to the CCN1-3 IGFBP domains although this is likely due to incomplete modelling rather than a biological difference. This figure was adapted from Holbourn et al (2008) with permission from the authors
The CCN1, 2, 3 and 6 IGFBP domains were successfully modelled by the CPH model server (Lund et al. 2002), (Figs. 3, 4, 5 and 6). Poor sequence similarity between the IGFBP4 template and CCN4 and CCN5 are the likely reason why the program could not model the domain for all CCN proteins. Even then, the model of CCN6 is poor missing out many of the features but this is likely due to it being an incomplete model rather than any significant biological difference. The models of CCN1-3 when compared to IGFBPs maintain the same overall structure with parallel loops supported by a ladder of 3 disulphide bonds and the flat palm and globular finger regions flanking the IGF binding cleft. Despite the high degree of similarity between the models and the known IGFBP structures there are some significant differences in key areas, most importantly the thumb region and the IGF binding cleft. The thumb region is missing in the CCN family. In the traditional IGFBPs the thumb has a conserved XhhyC motif (‘h’ is a hydrophobic amino acid and ‘y’ is positively charged residue) (Kalus et al. 1998; Siwanowicz et al. 2005; Sitar et al. 2006) but as can be seen in the sequence alignment in Fig. 1 the CCN molecules have a wide range of different amino acids in this region. The second difference lies in IGF binding cleft which in IGFBP4 and 5 is comprised mainly of hydrophobic residues, including in the case of IGFBP5 a short leucine rich segment (Kalus et al. 1998). In the CCN proteins this section is a mix of different amino acids although the effect that this variation has is unknown, but may go some way to explaining the weak to non-existent IGF binding displayed by the CCN family. When the electrostatic surfaces of each molecule are displayed other differences could be observed. Whist the molecules share the same overall structure and disulphide bonding patterns the changes in surface properties may begin to account for the differences in binding partners and activities between the members of the CCN family.
The von Willebrand factor C repeat domain
The von Willebrand factor type C (VWC), also known as the chordin-like cysteine rich (CR) repeat, is found in >500 extracellular matrix proteins making it one of the most common domains found in the genome (Zhang et al. 2007). The molecules that it can be found in are varied and include CCN proteins, procollagen, thrombospondin, von Willebrand factor, glycosylated mucins and neuralins (Abreu et al. 2002). In many of these proteins there are multiple copies of the VWC domain. For example the von Willebrand factor (Mancuso et al. 1989) contains 2 repeats or chordin that contains 4 repeats (O’Leary et al. 2004). The CCN proteins are perhaps slightly unusual in that it has a single copy, and in the case of CCN6 it is an incomplete copy lacking four of the 10 conserved cysteine residues (Bork 1993).
Regulation of bone morphogenic proteins (BMPs) and transforming growth factor beta (TGF-β) are the two most common functions ascribed to the VWC domain and common across many proteins it can be found in (Zhu et al. 1999; Nakayama et al. 2001; Sakuta et al. 2001; Abreu et al. 2002). Both TGF-β and the BMPs are important members of a large family of small growth factors such as TGF-β, Vascular endothelial growth factor (VEGF) and Placenta growth factor (PlGF). In the case of TGF-β and the BMPs they are regulating molecules responsible for organ growth and development for the kidneys, lungs and teeth as well as in skeletal formation, patterning and influencing the growth of both bone and cartilage (Hogan 1996a; Hogan 1996b).
The interaction between the CCN proteins and the small growth factor family is a key one that is responsible for many of the CCN protein functions and involves both the VWC and TSP domains. Some important functions that the CCN-growth factor interactions regular include: TGF-β mediated adhesion and tissue remodelling (Perbal and Takigawa 2005), induction of angiogenesis (Lau and Lam 1999), kidney development (Joliot et al. 1992), chondrogenic and skeletal development (O’Brien and Lau 1992; Kireeva et al. 1996; Wong et al. 1997) and a host of other TGF-β related pathways (Brigstock 1999; Lau and Lam 1999; Perbal 2004).
The range of substrate specificity for these growth factors across the spectrum of VWC domains found in humans is quite large. In addition, given the many varied biological functions these repeats have, the differences in substrate or relative strengths of binding substrate binding are likely to play a role in the VWC domain’s biological role. For example CCN2 binds to BMP4 and TGF- β1, (Abreu et al. 2002) and CCN3 binds to BMP2 (Minamizato et al. 2007) whilst chordin binds to BMP-4, −5 and −6 as well as TGF-β1 and -β2 (Nakayama et al. 2001) despite the high similarity between the VWC domains. In addition while CCN2 binds both TGF-β1 and BMP4 it has a higher affinity for BMP4 (KD 5nM compared with 30nM for TGF-β1) (Abreu et al. 2002) which may be important for the role of CCN2 in BMP and TGF-β related functions. In the case of CCN2 the relatively low affinity for TGF-β1 may explain CCN2's role as a chaperone of TGF-β1 transferring it between high affinity receptors with affinities in the picomolar range (Massague 1987) rather than accentuating TGF-β1 signalling. By comparison the strong interaction with BMP4 results in inhibition of BMP4’s activity (Abreu et al. 2002).
A second major function of the CCN family of proteins is in modulating the contents of the ECM; and interactions between the VWC domain and growth factors may hold the key to this. Several important ECM molecules such as collagen and fibronectin can be modulated by the CCN proteins and may take place through induction via TGF-β1 (Roberts et al. 1986; Brigstock 1999; Abreu et al. 2002). Recently, the physical interaction of CCN3 and BMP2 was shown to inhibit BMP2-induced osteoblast differentiation (Minamizato et al. 2007). Interactions between CCN molecules and growth factors may also play roles in tumour formation and cell development as with some CCN3 biological mutants lacking the VWC domain being found in both Wilms tumours and in Ewing’s tumours (Perbal et al. unpublished results). Similarly CCN4 lacking the VWC domain has been linked to schirrhous gastric carcinoma (Tanaka et al 2001).
Lastly, a putative third function for the VWC domain, in addition to interacting directly with growth factors and involvement in cancer, for the VWC domain may be in large scale oligomerisation of CCN molecules. Its is known that in von Willebrand Factor it is the VWC domain that is responsible for forming large scale oligomers after a separate dimerisation step has taken place, and it is possible that this also be the case in CCN molecules. The CT domain is well known for its ability to form both homo- and hetero-dimers of CCN molecules (Perbal et al. 1999) and if the VWC domain can then make larger oligomers of homo- and hetero- nature this may add an additional layer of complexity to CCN regulation (Voorberg et al. 1991; Brigstock 1999; Perbal 2001b). Another reason for the oligomerisation may come about through an additive affinity of multiple VWC domains working in concert. In chordin and the other multiple copy proteins the affinity for growth factors varies between repeats and the activity of intact proteins with multiple VWC domains, can be as much as 10 fold higher than that for an individual repeat suggesting a complicated means of regulation (Larrain et al. 2000). The significance of this effect upon the function and regulatory activity of the single VWC domain in CCN proteins is not yet known.
The VWC repeat is a short ~70 amino acid stretch containing 10 cysteine residues that are part of the two motifs that were used to classify this domain (Bork 1993). These motifs are characterised by the cysteine pattern and numbered as such (with the number referring to which of the 10 conserved cysteines is). The first: C2xxC3xC4, lies towards the middle of the repeat and the second, C8C9xxC10, lies towards the end of the repeat. These motifs are conserved in the CCN proteins as can be seen in Fig. 1 but with slight modifications. Both are extended with an extra residue between C2 and C3 and two extra residues between C9 and C10 (Bork 1993) except in CCN6 where 4 cysteine residues are missing (numbers 2, 6, 8 and 9) (Bork 1993).
The structures of several different VWC domains are known including those from collagen [PDB IU5M] (O’Leary et al. 2004) determined by NMR and the chordin family member crossveinless 2 (CV2) (from Drosophila) [PDB 3BK3] (Zhang et al. 2008) which was determined by X-ray crystallography. Both domains, as would be expected, possess a highly similar structure. The domain, shown in Fig. 2, forms an extended “boot-like” shape made up of two roughly equal size sub domains- subdomain 1 (SD1) is more structured and comprises the N-terminal part of the domain with a short two stranded anti-parallel β-sheet followed by a 3 stranded anti-parallel β-sheet. The triple sheet is supported by a disulphide bond between strands 2 and 3, and a second disulphide bridge formed between strand 2 and the first strand of the two-stranded sheet. The second sub-domain (SD2) is far less structured being comprised of random coil with no secondary structure elements but is constrained by 3 disulphide bridges into a novel fold that is reminiscent in fibronectin (O'Leary et al. 2004; Zhang et al. 2008) and may possess similar interactions with growth factors (O'Leary et al. 2004). An additional important feature is also present in the Drosophila CV2 structure at the extreme N-terminal region formed from the first 8 or so residues that has been called the “clip” domain as it wraps around the BMP substrate like a paperclip with several hydrogen bonds; although this clip region is not seen in the VWC domain from chordin (Zhang et al. 2008; O’Leary et al. 2004). In BMPs, TGF-β1 and the related growth factors there are two binding epitopes that have been recognised (Zhang et al. 2007), a “knuckle” and “wrist” region that are crucial for receptor binding. There are some results that suggest in the case of the VWC repeats' antagonistic behaviour towards BMPs that they interfere with the interactions between the “knuckle epitope” and the BMP-receptor II (Keller et al. 2004; O'Leary et al. 2004). Some of the biological effects of the VWC domains may come about from the domain blocking access to these epitopes and inhibiting receptor binding. In the case of interaction between BMPs and a variety of VWC domains it has become clear there are two main modes of binding. Those that bind at the “wrist” epitope involve mainly hydrophobic interactions and those that bind at the “knuckle” epitope that have hydrophobic interactions and the “clip” region that acts as an “affinity enhancer” through a series of hydrogen bonds (Zhang et al. 2007; Zhang et al. 2008). In the structure of BMP2 bound to Drosophila CV2, it was strong hydrophobic interactions on a relatively small area of the face of SD1 and hydrogen bonds from the clip region that were responsible for high affinity binding (Zhang et al. 2008).
Models of the six CCN VWC domains were generated using the CPH model server (Lund et al. 2002) and NMR structure of the VWC-like domain from collagen IIA [PDB IU5M] (O'Leary et al. 2004) as a template and can be seen in Figs. 3, 4, 5 and 6. The only model that did not have all 10 cysteines visible, including those present in the two conserved motifs (C2xxC3xC4; C8C9xxC10) was CCN2 that was slightly truncated but this is due to the modelling program rather than any significant differences in the CCN2 VWC domain. All of the modelled domains share the same two sub-domain layout with a more structured sub-domain at the N-terminal end and a fibronectin-like fold of random coil supported by the disulphide bridges at the C-terminal end (O'Leary et al. 2004; Zhang et al. 2008). Also like the chordin structure [PDB 1U5M](O'Leary et al. 2004), they do not possess the clip region that was seen in the CV2 structure [PDB 3BK3](Zhang et al. 2008). As the position of this clip region in amino acid sequence is located in the short linker region between the IGFBP and VWC domains it is likely that it is not present in the CCN VWC domain. In the case of the interactions between TGF-β and the variety of BMPs that are known to bind CCN molecules it is not possible to speculate on the exact nature of the interaction or whereabouts on the VWC domain that this interaction takes place though the lack of space in the sequence to form a “clip” segment would suggest that it would bind the wrist epitope of the BMPs (Zhang et al. 2008). Furthermore, while CCN proteins closely resemble each other in their arrangement a look at their electrostatic surfaces, shown in Figs. 3, 4, 5 and 6, illustrates a wide range of differences between them. CCN1 and to a lesser extent CCN5 are primarily negatively charged on the front face of the VWC domain whilst CCN4 is primarily positively charged. The remaining molecules have a mix of charges on their surface. The large differences in surface charge may play a part in the different behaviours of CCN family members or how they can arrange themselves with either inter- or intra- molecular oligomerisation. A second reason for the difference between the family members may be in differences in the loop regions surrounding the disulphide core as this method of substrate selectivity has been observed in the disulphide rich conotoxins (Zhang et al. 2007; Armishaw and Alewood 2005).
The thrombospondin type 1 (TSP) domain
The TSP domain is a short sequence (~55 residues) that is found in 187 TSP proteins within the human genome and numerous other eukaryotic organisms (Tucker 2004). There is a large number of mostly extracellular matrix associated proteins that possess a TSP domain and these include: thrombospondins and spondins, papilin, extracellular matrix ADAMTS, mindin and complement pathway proteins (C6, C7, C8A, C8B, C9 and properdin) (Adams and Tucker 2000; Tucker 2004). The domain is named after the thrombospondin family of angiogenic regulators. This family members all share a common structure including a three type I thrombospondin repeats (TSP domain), three epidermal growth factor-like repeats (thrombospondin type -2 repeats) and seven aspartic acid rich repeats (thrombospondin type-3 repeats) (Tucker 2004; Lawler and Hynes 1986; Iruela-Arispe et al. 2004; Karagiannis and Popel 2007).
From studies on thrombospondin and other TSP containing proteins the domain seems to have 4 common functions- a) cell attachment sites in signalling and adhesion, b) regulation of angiogenesis, c) protein binding sites for a range of growth factors and other ECM proteins and d) glycosaminoglycan (GAG) binding sites (Chen et al. 2000). These functions obviously cover a wide range of biological roles and necessitate a diverse array of binding partners that can interact with the TSP domain. Many of these have been identified and include key signalling molecules such as collagen V (Takagi et al. 1993), fibronectin (Sipes et al. 1993), CD36 (Asch et al. 1992), TGF-B (Schultz-Cherry et al. 1995) and heparin (Guo et al. 1992) and a wide range of extracellular proteins.
The TSP domain in the CCN proteins is known to have a strong role in adhesion and modulation of ECM proteins (Planque and Perbal 2003) through interactions with the lipoprotein- related receptor (Heng et al. 2006; de Winter et al. 2008), binding to sulphated glycoconjugates (Holt et al. 1990) and interacting with several different integrins (Kubota and Takigawa 2007).
Thrombospondin and related proteins are a well known and potent family of angiogenic regulators (Folkman 1996; Karagiannis and Popel 2007) as is the CCN family (Perbal 2004; Kubota and Takigawa 2007) and the TSP domain is an important component of their angiogenic property. In CCN proteins both the TSP domain and the CT domain interact with VEGF- one of the key angiogenic growth factors (Inoki et al. 2002). Whilst VEGF can be found in several isoforms the TSP domain of CCN2 binds to the heparin-binding VEGF165 isoform in an anti-angiogenic mode of action, whilst the CT domain is involved in interactions with both VEGF165 and VEGF121. The TSP domain of other CCN proteins has been confirmed to have an anti-angiogenic mode of action and indeed only an isolated fragment of the TSP domain was needed to inhibit proliferation and migration of HUVEC cells (Karagiannis and Popel 2007; Tong and Brigstock 2006). This anti-angiogenic effect can be removed by treating CCN2 with matrix metalloproteases (MMPs) that are known to target the spacer regions between the CCN modules (Ball et al. 1998; Inoki et al. 2002) suggesting that anti-angiogenic effect comes about from the TSP binding to and sequestering the VEGF165 away from its receptors and the MMP cleavage frees the VEGF. Another small growth factor that interacts with the TSP domain is TGF-β. In other thrombospondins such as thrombospondin -1, this requires an RFK tripeptide sequence found between the 1st and 2nd TSP domains of the protein. However, the TSP-1 domains in all of the CCN proteins lack this sequence so it may be another module is involved with TGF-β binding (Tan et al. 2002) or a combination of the TSP domain working synergistically with another of the TGF-β binding domains such as the CT domain that contains a reversed KFR tripeptide sequence (Kubota et al. 2006).
In addition to modulating angiogenesis through interactions with VEGF the TSP domain may have other biological functions mediated by integrin binding. Integrin α6β1 is a key receptor for TSP and responsible for many of the biological effects determined by the TSP domains (Tong and Brigstock 2006). Both CCN1 and CCN2 promote adhesion of fibroblasts and vascular smooth muscle cells through interactions with integrins and heparin sulphated proteoglycans with which the TSP domain is known to interact in support this hypothesis (Adams and Tucker 2000). Signalling through integrin binding is thought to be essential for the activity of the CCN family (Lau and Lam 1999) and hence the presence of TSP1 domain may be vital to function of these proteins. TSP domains have also been shown to interact with TGF-β (Adams and Tucker 2000) and interactions with TGB-β are central to many of the roles that the CCN proteins play and its possible that the interactions with TGF-β may be co-ordinated by the TSP domain working in conjunction with other domains of the CCN protein (Brigstock 1999; Lau and Lam 1999; Brigstock 2003). Like other domains it is possible that the TSP domain has some involvement in cancers (Perbal 2006) and there have been some studies that have linked CCN proteins with mutated or missing TSP domains with colorectal and gastric carcinomas (Thorstensen et al. 2001; Tanaka et al. 2002) as well as Wilm’s tumours (Subramaniam et al. 2008).
The TSP domain in the CCN proteins contains the motifs found in many other TSP repeats throughout the eukaryotic genomes. As it is so common several TSP domains have had their structures determined through a combination of both X-ray crystallography and NMR. These structures include: thrombospondin (Tan et al. 2002), malaria TRAP protein (Tossavainen et al. 2006) and F-spondin (Paakkonen et al. 2006). Using the CPH modelling server (Lund et al. 2002) the structures of malaria TRAP protein [PDB ID 2BBX] (Tossavainen et al. 2006) (for CCN 1, 3 and 5) or thrombospondin-1 [PDB ID 1LSL] (Tan et al. 2002) (for CCN 2,4 and 6) were used to construct models of the TSP domains for all six CCN proteins. In each case the models were about 45 residues in length and contained the same structural pattern as that of the actual thrombospondin structures and are shown in Figs. 3, 4, 5 and 6. The TSP domain is a small ~55 residue domain comprised of a small 3-stranded anti-parallel β-sheet (approximately 15 × 20 × 55 Å) that is twisted slightly into a right handed helical shape where the first strand is a more irregular and resembles random coil but still maintains hydrogen bonds with the adjacent strand (Tan et al. 2002). There are 6 conserved cysteines that are all used in inter-domain disulphide bridges and a conserved CSxTCG motif, which can be seen in the CCN proteins in Fig. 1 and the ribbon diagram in Fig. 2 (except in CCN3 that substitutes the T for an S). The other important structural feature of the TSP domains is the “CWR” layers. These are an array of hydrogen bonds between residues that form the faces of the β-strands and alongside the disulphide bonds give rise to a ladder-like series of bonded amino acids. Each layer is named from the amino acid that forms the hydrogen bonds; cysteine (C), tryptophan (W) or arginine (R). These layers in the other thrombospondin containing proteins come about from a strongly conserved WxxWxxW motif and a pair of well conserved arginines (Bork 1993; Tan et al. 2002). The 3 conserved disulphide bonds present in each TSP domain link the turns together at the top and bottom of the sheet to stabilise the structure. The complete series of 3 disulphide bridges can only be seen in the model of CCN5 as the other models are slightly truncated due to the limitations of structural homology modelling. But if the models could be extended then all of the cysteines are conserved in the protein sequence to form the correct sulphur-sulphur bonds. Based on the pattern of these disulphide bridges and the strands where the cysteines appear, Tan et al (2002) broadly divided the TSP family into two broad groups. In the models of the CCN TSP domains the disulphide pattern would place the CCN domains in group 2 alongside F-Spondin, TRAP and the various proteins of the complement system (Tan et al. 2002). However the domains in the CCN family lack the tryptophan rich motif (only possessing a single tryptophan residue) and only having a single arginine residue leading to less CWR layers (Figs. 3, 4, 5 and 6). The importance of the CWR layers beyond maintaining the structure of the domain is unknown as the biologically active section has been determined to be a short section of ~12 residues that makes up the first strand and part of the second (Karagiannis and Popel 2007; Tong and Brigstock 2006). These short peptides have been shown to be biologically active for CCN1–3, 5 and 6 (Karagiannis and Popel 2007). In addition to the short biologically active section of strands 1 and 2 there is another unifying feature of the TSP domain shared between all 6 members of the CCN family. This is a groove running across the face of the molecule that is lined with basic residues and is large enough to accommodate 2 heparin molecules. The basic residues would be available to form strong electrostatic bonds with negatively charged sulphur groups on sulphated heparin molecules. As this is conserved in all six of the models it might suggest that all the CCN proteins use the TSP domain in a similar manner for binding to heparin or sulphated proteoglycans to modulate cell adhesion and ECM composition in a similar manner to thrombospondin and other TSP containing proteins (Tan et al. 2002).
The cysteine knot C-terminal (CT) domain
The CT domain is named after the ‘cysteine knot’ motif that it contains (Bork 1993). Cysteine knots are common to many other proteins both large mosaic extracellular matrix proteins such as Drosophila slit protein, the von Willebrand factor, several mucins, Norrie disease protein and the TGF-β family of small growth factors (Bork 1993; McDonald and Hendrickson 1993). This family includes TGF, VEGF, BMPs, nerve growth factor (NGF), platelet derived growth factor (PDGF) and the Norrie disease protein (Bork 1993; McDonald and Hendrickson 1993).
Many of the biological functions of the small growth factors come about through interactions with heparin and heparin sulphate proteoglycans (HSPGs) (Lyon et al. 1997; Rider 2006). While the TSP domain is also thought to bind heparin or HSPGs it has been confirmed that the CT domain can do the same, porcine CCN2 fragments consisting of just the ~10 kDa CT domain bind heparin strongly and are biologically active (Brigstock et al. 1997). In the CCN proteins the interactions with heparin and HSPGs are likely connected to many of the important roles in controlling and manipulating adhesion processes and the composition of the ECM alongside. It is well known that heparin is an important modulator in adhesion processes and in forming and controlling the extracellular matrix. Interactions between the CT domain and heparin are not the only method through which CCN molecules can control adhesion. Interactions between the CT domain and Fibulin 1C (Perbal et al. 1999), and several different integrins such as αVβ5, αvβ3, αmβ2 and α5β1 (Grzeszkiewicz et al. 2001; Gao and Brigstock 2004; Gao and Brigstock 2006) have been observed and that the constructs containing only the CT domain can bring about cell adhesion (Ball et al. 2003). The large number of integrins that interact with the CT domain do so through several different binding sites although in the CCN proteins none of these use the common RGD tripeptide - integrin binding motif seen in many other proteins. Although interestingly in CCN2 integrin α5β1 binds to a DGR motif, possibly a reverse integrin binding site (Gao and Brigstock 2006).
A second role for the CT domain in regulating mitogenic effects cell differentiation and other mitogenic processes may come about through interactions with cell differentiation molecule Notch 1 (Sakamoto et al. 2002) and the apoptosis inducing integrin α6β1 (Todorovicc et al. 2005) and artifical truncates of the CT domain have been seen to promote and control these effects (Brigstock et al. 1997). In addition to its mitogenic effects the isolated CT domain of CCN3 has been seen to possess a anti-proliferative function preventing both proliferation and differentiation of mesenchymal stem cells (Katsuki et al. 2008) and was recently reported to contain sequences that are involved in the nuclear addressing of CCN variants lacking the signal peptide (Planque et al. 2006).
While the sheer number of pathways that can be regulated and controlled by HSPGs may lead many to consider HSPGs and integrins as the “functional receptors for the CCN family” (Leask and Abraham 2006). There are additional functions independent of the integrin/HSPG pathways. Recombinant constructs of the CT domain can modulate the Wnt pathway through interactions with the LDL receptor protein 6 (LRP6) (Mercurio et al. 2004; Latinkic et al. 2003). Though as the Wnt relies on integrins to recruit some the associated proteins it is possible that the modulation of the Wnt pathway comes about from convergent actions of the CT domain on Wnt complex and integrins (Lau and Lam 1999; Marsden and DeSimone 2001; Mercurio et al. 2004). All of these roles suggest that the part played by the CT domain in CCN function is highly important and as yet still not fully understood.
The cysteine knot containing ECM proteins and small growth factors all share a cysteine knot motif though the range of molecules possessing this motif exhibit significant sequence variation outside of the core knot fold (McDonald and Hendrickson 1993). A cysteine knot is an 8 residue ring based around a two-stranded anti-parallel β-sheet (with each strand at least 4 residues long) linked by 2 disulphide bonds to complete the ring and a third bond through the centre of the “knot” (Fig. 2). In some cases there is a short alpha-helix on the opposite side of the cysteine knot rather than two β-strands (McDonald and Hendrickson 1993; Schlunegger and Grutter 1993; Rider 2006; Isaacs 1995). Many of the growth factors are found naturally as dimers linked through disulphides. In the case of both NGF and TGF-β the disulphide that threads through the centre of the knot is inter-chain and it is a different cysteine that is responsible for the dimer formation, whilst in platelet derived growth factor (PDGF) it is the cysteine passing through the centre of the knot that forms an intra-chain disulphide bond to complete the dimer. It is unknown in the CCN proteins which of these arrangements the CT domain follows although it is the CT domain responsible for forming CCN dimers (Brigstock 1999; Perbal et al. 1999). Using the CT domain of CCN proteins both homo- and hetero-dimers can be formed as the CT domain of CCN3 has been seen to interact with CCN2 in GST pull-down assays (Perbal et al. 1999). It is possible that this dimerisation about the CT domains is followed by oligomerisation about the VWC domain to produce the CCN oligomers that have been observed (Planque and Perbal 2003).
The sequence diversity outside of the core cysteine knot motif made modelling the CT domains of the CCN proteins more difficult. The CPH model server (Lund et al. 2002) was unable to construct models of the CT domain for any of the CCN molecules. Instead the Phyre homology and recognition server was used (Kelley et al. 2000; Bennett-Lovsey et al. 2008). Even then the Phyre server was only able to build partial models for CCN1-3. These partial models contained ~50 residues out of the ~80 residue domain though this did include the cysteine knot. Due to sequence variation a different template had to be used in each case. The BMP7 was the template structure for CCN1 [PDB ID 1LXI] (Greenwald et al. 2003), the TGF-β3 structure [PDB ID 1KTZ] (Hart et al. 2002) for CCN2 and TGF-β1 as a template [PDB ID 1KLA] (Hinck et al. 1996) for CCN3. The models can be viewed in Figs. 3, 4 and 5. In the visible sections of the models the cysteine knot can be seen formed by a two stranded anti-parallel β-sheet with the two disulphides that connect the β-strands and close the ring. A fifth cysteine protruding through the ring becomes available for a third disulphide bond formation. Though without a complete structure it is still unknown whether this third disulphide will be intra-molecular within the CT domain or inter-molecular and bind to other cysteine knot containing growth factors or form a dimer with other CCN molecules (Perbal et al. 1999). The heparin binding site in most growth factors is isolated at the tips of loops between the β-sheets and has 4 basic residues on the first sheet and a single Arg/Lys residue on the tip of loop on the second sheet (Lyon et al. 1997; Rider 2006). All of the CCN proteins except for CCN5 (that lacks the CT domain) possess a high number of basic residues at the N-terminal of the CT domain that follow the general heparin binding pattern of xBBxBx (where B is a basic residue and x is usually not charged) (Cardin and Weintraub 1989). However the N-terminal residues including the heparin binding basic region and the loops at the end of the β-strands were unable to be modelled for any of the CCNs. While all of the CT domains seem to have a similar arrangement, the electrostatic surfaces of each appear to show some differences and this coupled with a fairly diverse amino acid sequence (apart from the conserved cysteines) may account for the wide range of ligands and binding partners that have been found for the CT domain. However, many of these differences may only become apparent with the availability of experimentally determined structures.
The CT domain with its heparin (Bork 1993; Brigstock et al. 1997), growth factor-like(Bork 1993), integrin and Wnt pathway binding (Mercurio et al. 2004) seems like it could be the major force for direct protein-protein interactions and the availability or presence of the other domains modulate the results of these interactions. The CT domain’s involvement with integrin and HSPG mediated pathways and mitogenic effects has been seen and biologically active truncates of the CT domain alone are enough to produce these effects and in CCN3 inhibit growth (Brigstock et al. 1997; Bleau et al 2007). This makes the CT domain one of the most important domains for CCN protein behaviour as it seems to govern many crucial aspects of CCN biology including dimerisation and also begs the question- “how does CCN5 manage many of the same effects of the other family members without the CT domain?”
Conclusions
CCN proteins all share the similar basic modular structure and a high degree of amino acid sequence similarity and play a part in many cellular functions. However the sheer number of functions and often opposing functions of different CCN molecules make it unlikely that these functions are wholly based upon the action of individual domains, but rather on a cumulative effect between multiple domains and receptors (Brigstock 1999; Lau and Lam 1999; Perbal 2001b). This is supported by the work performed by Kubota el al (Kubota et al. 2006) where many effects could be elicited by individual domains, in some cases elicited by all 4 domains, but other effects required the full length protein or a mixture of individual domains (Kubota et al. 2006).
The use of a small library of discrete domains acting as a building blocks to create much larger complex multifunctional extracellular associated proteins is a common occurrence in many eukaryotes (Hohenester and Engel 2002) and places the CCN proteins in the same category as many other multifunctional matrix associated proteins. However while the knowledge of which domains are present is useful it does not offer a straightforward explanation into how the modular structure of these proteins is able to control or regulate the complex behaviour of these proteins. Evidence of internal protein regulation by interactions between the domains is hard to obtain but the change in biological activity of truncates missing various domains would certainly support this idea. The flexible nature of the full length molecule with the long central linker region and shorter inter-domain linkers would also allow a great deal of flexibility within the CCN molecules. Also the nature of multiple domains binding to the same target, such as both domains III and IV binding to VEGF (Inoki et al. 2002) may indicate some synergy between domains in substrate binding, or an altering of subtle biological signals depending on the various mix of domains involved. In addition, the role of the CT as a dimerisation domain (Bork 1993) and the suspected role of the VWC domain in forming larger higher order oligomers may result in some of the functions being modulated by multiple CCN proteins in a larger complex working together (Bork 1993; Brigstock 1999; Perbal et al. 1999). In an oligomer multiple domains may act together upon substrate binding to mimic the way the VWC or TSP repeats function in other large multi-domain proteins (Adams and Tucker 2000; Mancuso et al. 1989). This is further complicated by the ability of domain IV to form heterodimers between differing CCN family members adding an additional layer of complexity as it is possible that some effects come about through a cumulative effect of different CCN molecules in a larger complex (Perbal et al. 1999).
The varied and often conflicting behaviour of the CCN family members may not just be down to subtle differences in structure but to other factors like proteolytic processing, control of gene expression or even the organ or spatial locations in the cell that each are expressed. This may suggest that time dependant expression of the CCN proteins, such as CCN1 and CCN2 being immediate early genes whilst CCN3 is not (Bradham et al. 1991; O’Brien and Lau 1992; Joliot et al. 1992) is an important functional level of regulation. The protease cleavage of the linker regions leads to differing functions due to the loss of certain domains in biologically isolated truncates (Tong and Brigstock 2006; Brigstock 1999) such as CCN2 losing its anti-angiogenic effects after treatment with certain MMPs (Hashimoto et al. 2002). The use of different expression regulatory systems and tissue specificities could go some way to explaining the conflicting range of functions that can be found amongst the CCN molecules despite the high degree of similarity at the amino acid sequence level (Brigstock 1999; Perbal 2001a; Perbal 2004) .
While approximate structures of each domain could be modelled based on their similarity (homology) with known structures, it is likely that the CCN proteins have some features that help explain their specificity and possibly a route through which they regulate themselves through inter-domain interactions. These could take place either through flexibility about the hinge regions or additional loops but these are features that would require structural determination of the proteins in question to confirm. Linker regions acting as “hinges” allowing large domain movements have been shown to be involved in modulation of activity of other mosaic proteins (Dobson 1990) and the nature of the linker regions between domains of large ECM proteins, be they helical or elongated, may also play a role (Arai et al. 2004). The ability of linker regions to be used as means of inter-domain communication in large molecules may also prove important in given the large variation in sequence and length present in the CCN protein family (Bork 1993; Brigstock 1999; Gokhale and Khosla 2000). The susceptibility of the linker to proteolytic cleavage and biologically active nature of many truncated CCN constructs may lead to another layer of physiological control of CCN function (Bork 1993; Brigstock et al. 1997; Ball et al. 1998). However the flexible nature of the linkers means that they can not be modelled. Hence it is hard to speculate on the effect that differences in the linkers will have.
The differences in sequence can lead to large variations in surface charge and amino acid composition and currently unknown active sites whilst still retaining the same core shape. The nature of the long hinge region and the observed flexible natures of the VWC and TSP domains from their related structures (Tan et al. 2002; O'Leary et al. 2004) may also allow the N and C terminal of the CCN proteins to interact with each other as suggested by Perbal (Perbal 2001b). The determination of the structure of each domain; either individually or full length protein is necessary in order to answer some of the key questions about the CCN family of proteins by correlating structure to function (Perbal 2001a).
References
Abreu JG, Ketpura NI, Reversade B, De Robertis EM (2002) Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-beta. Nat Cell Biol 4:599–604
Adams JC, Tucker RP (2000) The thrombospondin type 1 repeat (TSR) superfamily: diverse proteins with related roles in neuronal development. Dev Dyn 218:280–299. doi:10.1002/(SICI)1097-0177(200006) 218:2<280::AID-DVDY4>3.0.CO;2-0
Arai R, Wriggers W, Nishikawa Y, Nagamune T, Fujisawa T (2004) Conformations of variably linked chimeric proteins evaluated by synchrotron X-ray small-angle scattering. Proteins 57:829–838. doi:10.1002/prot.20244
Armishaw CJ, Alewood PF (2005) Conotoxins as research tools and drug leads. Curr Protein Pept Sci 6:221–240. doi:10.2174/1389203054065437
Asch AS, Silbiger S, Heimer E, Nachman RL (1992) Thrombospondin sequence motif (CSVTCG) is responsible for CD36 binding. Biochem Biophys Res Commun 182:1208–1217. doi:10.1016/0006-291X(92)91860-S
Ball DK, Surveyor GA, Diehl JR, Steffen CL, Uzumcu M, Mirando MA, Brigstock DR (1998) Characterization of 16- to 20-kilodalton (kDa) connective tissue growth factors (CTGFs) and demonstration of proteolytic activity for 38-kDa CTGF in pig uterine luminal flushings. Biol Reprod 59:828–835. doi:10.1095/biolreprod59.4.828
Ball DK, Rachfal AW, Kemper SA, Brigstock DR (2003) The heparin-binding 10 kDa fragment of connective tissue growth factor (CTGF) containing module 4 alone stimulates cell adhesion. J Endocrinol 176:R1–R7. doi:10.1677/joe.0.176R001
Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA (2008) Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins 70:611–625. doi:10.1002/prot.21688
Bleau AM, Planque N, Lazar N, Zambelli D, Ori A, Quan T, Fisher G, Scotlandi K, Perbal B (2007) Antiproliferative activity of CCN3: involvement of the C-terminal module and post-translational regulation. J Cell Biochem 101:1475–1491. doi:10.1002/jcb.21262
Bork P (1993) The modular architecture of a new family of growth regulators related to connective tissue growth factor. FEBS Lett 327:125–130. doi:10.1016/0014-5793(93)80155-N
Bornstein P (1995) Diversity of function is inherent in matricellular proteins: an appraisal of thrombospondin 1. J Cell Biol 130:503–506. doi:10.1083/jcb.130.3.503
Bradham DM, Igarashi A, Potter RL, Grotendorst GR (1991) Connective tissue growth factor: a cysteine-rich mitogen secreted by human vascular endothelial cells is related to the SRC-induced immediate early gene product CEF-10. J Cell Biol 114:1285–1294. doi:10.1083/jcb.114.6.1285
Brigstock DR (1999) The connective tissue growth factor/cysteine-rich 61/nephroblastoma overexpressed (CCN) family. Endocr Rev 20:189–206. doi:10.1210/er.20.2.189
Brigstock DR (2003) The CCN family: a new stimulus package. J Endocrinol 178:169–175. doi:10.1677/joe.0.1780169
Brigstock DR, Steffen CL, Kim GY, Vegunta RK, Diehl JR, Harding PA (1997) Purification and characterization of novel heparin-binding growth factors in uterine secretory fluids. Identification as heparin-regulated Mr 10,000 forms of connective tissue growth factor. J Biol Chem 272:20275–20282. doi:10.1074/jbc.272.32.20275
Brigstock DR, Goldschmeding R, Katsube KI, Lam SC, Lau LF, Lyons K, Naus C, Perbal B, Riser B, Takigawa M, Yeger H (2003) Proposal for a unified CCN nomenclature. Mol Pathol 56:127–128. doi:10.1136/mp. 56.2.127
Cardin AD, Weintraub HJ (1989) Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis 9:21–32
Chen H, Herndon ME, Lawler J (2000) The cell biology of thrombospondin-1. Matrix Biol 19:597–614. doi:10.1016/S0945-053X(00)00107-4
Davies SR, Watkins G, Mansel RE, Jiang WG (2007) Differential expression and prognostic implications of the CCN family members WISP-1, WISP-2, and WISP-3 in human breast cancer. Ann Surg Oncol 14:1909–1918. doi:10.1245/s10434-007-9376-x
de Winter P, Leoni P, Abraham D (2008) Connective tissue growth factor: structure-function relationships of a mosaic, multifunctional protein. Growth Factors 26:80–91. doi:10.1080/08977190802025602
Dobson CM (1990) Protein conformation. Hinge-bending and folding. Nature 348:198–199. doi:10.1038/348198a0
Firth SM, Baxter RC (2002) Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev 23:824–854. doi:10.1210/er.2001-0033
Folkman J (1996) Tumor angiogenesis and tissue factor. Nat Med 2:167–168. doi:10.1038/nm0296-167
Gao R, Brigstock DR (2004) Connective tissue growth factor (CCN2) induces adhesion of rat activated hepatic stellate cells by binding of its C-terminal domain to integrin alpha(v) beta(3) and heparan sulfate proteoglycan. J Biol Chem 279:8848–8855. doi:10.1074/jbc.M313204200
Gao R, Brigstock DR (2006) A novel integrin alpha5beta1 binding domain in module 4 of connective tissue growth factor (CCN2/CTGF) promotes adhesion and migration of activated pancreatic stellate cells. Gut 55:856–862. doi:10.1136/gut.2005.079178
Gokhale RS, Khosla C (2000) Role of linkers in communication between protein modules. Curr Opin Chem Biol 4:22–27. doi:10.1016/S1367-5931(99)00046-0
Greenwald J, Groppe J, Gray P, Wiater E, Kwiatkowski W, Vale W, Choe S (2003) The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly. Mol Cell 11:605–617. doi:10.1016/S1097-2765(03)00094-7
Grzeszkiewicz TM, Kirschling DJ, Chen N, Lau LF (2001) CYR61 stimulates human skin fibroblast migration through Integrin alpha vbeta 5 and enhances mitogenesis through integrin alpha vbeta 3, independent of its carboxyl-terminal domain. J Biol Chem 276:21943–21950. doi:10.1074/jbc.M100978200
Guo NH, Krutzsch HC, Negre E, Zabrenetzky VS, Roberts DD (1992) Heparin-binding peptides from the type I repeats of thrombospondin. Structural requirements for heparin binding and promotion of melanoma cell adhesion and chemotaxis. J Biol Chem 267:19349–19355
Hart PJ, Deep S, Taylor AB, Shu Z, Hinck CS, Hinck AP (2002) Crystal structure of the human TbetaR2 ectodomain-TGF-beta3 complex. Nat Struct Biol 9:203–208
Hashimoto G, Inoki I, Fujii Y, Aoki T, Ikeda E, Okada Y (2002) Matrix metalloproteinases cleave connective tissue growth factor and reactivate angiogenic activity of vascular endothelial growth factor 165. J Biol Chem 277:36288–36295. doi:10.1074/jbc.M201674200
Helle SI (2004) The insulin-like growth factor system in advanced breast cancer. Best Pract Res Clin Endocrinol Metab 18:67–79. doi:10.1016/S1521-690X(03)00045-9
Heng EC, Huang Y, Black SA Jr, Trackman PC (2006) CCN2, connective tissue growth factor, stimulates collagen deposition by gingival fibroblasts via module 3 and alpha6- and beta1 integrins. J Cell Biochem 98:409–420. doi:10.1002/jcb.20810
Hinck AP, Archer SJ, Qian SW, Roberts AB, Sporn MB, Weatherbee JA, Tsang ML, Lucas R, Zhang BL, Wenker J, Torchia DA (1996) Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2. Biochemistry 35:8517–8534. doi:10.1021/bi9604946
Hogan BL (1996a) Bone morphogenetic proteins in development. Curr Opin Genet Dev 6:432–438. doi:10.1016/S0959-437X(96)80064-5
Hogan BL (1996b) Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev 10:1580–1594. doi:10.1101/gad.10.13.1580
Hohenester E, Engel J (2002) Domain structure and organisation in extracellular matrix proteins. Matrix Biol 21:115–128. doi:10.1016/S0945-053X(01)00191-3
Holbourn KP, Acharya KR, Perbal B (2008) The CCN family of proteins: structure-function relationships. Trends Biochem Sci 33:461–473. doi:10.1016/j.tibs.2008.07.006
Holt GD, Pangburn MK, Ginsburg V (1990) Properdin binds to sulfatide [Gal(3-SO4) beta 1–1 Cer] and has a sequence homology with other proteins that bind sulfated glycoconjugates. J Biol Chem 265:2852–2855
Hwa V, Oh Y, Rosenfeld RG (1999) The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr Rev 20:761–787. doi:10.1210/er.20.6.761
Inoki I, Shiomi T, Hashimoto G, Enomoto H, Nakamura H, Makino K, Ikeda E, Takata S, Kobayashi K, Okada Y (2002) Connective tissue growth factor binds vascular endothelial growth factor (VEGF) and inhibits VEGF-induced angiogenesis. FASEB J 16:219–221
Iruela-Arispe ML, Luque A, Lee N (2004) Thrombospondin modules and angiogenesis. Int J Biochem Cell Biol 36:1070–1078. doi:10.1016/j.biocel.2004.01.025
Isaacs NW (1995) Cystine knots. Curr Opin Struct Biol 5:391–395. doi:10.1016/0959-440X(95)80102-2
Joliot V, Martinerie C, Dambrine G, Plassiart G, Brisac M, Crochet J, Perbal B (1992) Proviral rearrangements and overexpression of a new cellular gene (nov) in myeloblastosis-associated virus type 1-induced nephroblastomas. Mol Cell Biol 12:10–21
Jones JI, Clemmons DR (1995) Insulin-like growth factors and their binding proteins: biological actions. Endocr Rev 16:3–34
Kalus W, Zweckstetter M, Renner C, Sanchez Y, Georgescu J, Grol M, Demuth D, Schumacher R, Dony C, Lang K, Holak TA (1998) Structure of the IGF-binding domain of the insulin-like growth factor-binding protein-5 (IGFBP-5): implications for IGF and IGF-I receptor interactions. EMBO J 17:6558–6572. doi:10.1093/emboj/17.22.6558
Karagiannis ED, Popel AS (2007) Peptides derived from type I thrombospondin repeat-containing proteins of the CCN family inhibit proliferation and migration of endothelial cells. Int J Biochem Cell Biol. 39:2314–2323
Katsuki Y, Sakamoto K, Minamizato T, Makino H, Umezawa A, Ikeda MA, Perbal B, Amagasa T, Yamaguchi A, Katsube K (2008) Inhibitory effect of CT domain of CCN3/NOV on proliferation and differentiation of osteogenic mesenchymal stem cells, Kusa-A1. Biochem Biophys Res Commun 368:808–814. doi:10.1016/j.bbrc.2008.02.010
Keller S, Nickel J, Zhang JL, Sebald W, Mueller TD (2004) Molecular recognition of BMP-2 and BMP receptor IA. Nat Struct Mol Biol 11:481–488. doi:10.1038/nsmb756
Kelley LA, MacCallum RM, Sternberg MJ (2000) Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol 299:499–520. doi:10.1006/jmbi.2000.3741
Kim HS, Nagalla SR, Oh Y, Wilson E, Roberts CT Jr, Rosenfeld RG (1997) Identification of a family of low-affinity insulin-like growth factor binding proteins (IGFBPs): characterization of connective tissue growth factor as a member of the IGFBP superfamily. Proc Natl Acad Sci USA 94:12981–12986. doi:10.1073/pnas.94.24.12981
Kireeva ML, Mo FE, Yang GP, Lau LF (1996) Cyr61, a product of a growth factor-inducible immediate-early gene, promotes cell proliferation, migration, and adhesion. Mol Cell Biol 16:1326–1334
Kubota S, Takigawa M (2006) CCN2. In: Riley B., editor. AfCS-Nature Molecule Pages. http://www.signaling-gateway.org/
Kubota S, Takigawa M (2007) CCN family proteins and angiogenesis: from embryo to adulthood. Angiogenesis 10:1–11. doi:10.1007/s10456-006-9058-5
Kubota S, Kawaki H, Kondo S, Yosimichi G, Minato M, Nishida T, Hanagata H, Miyauchi A, Takigawa M (2006) Multiple activation of mitogen-activated protein kinases by purified independent CCN2 modules in vascular endothelial cells and chondrocytes in culture. Biochimie 88:1973–1981. doi:10.1016/j.biochi.2006.07.007
Larrain J, Bachiller D, Lu B, Agius E, Piccolo S, De Robertis EM (2000) BMP-binding modules in chordin: a model for signalling regulation in the extracellular space. Development 127:821–830
Latinkic BV, Mercurio S, Bennett B, Hirst EM, Xu Q, Lau LF, Mohun TJ, Smith JC (2003) Xenopus Cyr61 regulates gastrulation movements and modulates Wnt signalling. Development 130:2429–2441. doi:10.1242/dev.00449
Lau LF, Lam SC (1999) The CCN family of angiogenic regulators: the integrin connection. Exp Cell Res 248:44–57. doi:10.1006/excr.1999.4456
Lawler J, Hynes RO (1986) The structure of human thrombospondin, an adhesive glycoprotein with multiple calcium-binding sites and homologies with several different proteins. J Cell Biol 103:1635–1648. doi:10.1083/jcb.103.5.1635
Leask A, Abraham DJ (2006) All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J Cell Sci 119:4803–4810. doi:10.1242/jcs.03270
Lund O, Nielsen M, Lundegaard C, Worning P (2002). CPHmodels 2.0: X3M a Computer Program to Extract 3D Models. Abstract at the CASP5 conferenceA102.
Lyon M, Rushton G, Gallagher JT (1997) The interaction of the transforming growth factor-betas with heparin/heparan sulfate is isoform-specific. J Biol Chem 272:18000–18006. doi:10.1074/jbc.272.29.18000
Mancuso DJ, Tuley EA, Westfield LA, Worrall NK, Shelton-Inloes BB, Sorace JM, Alevy YG, Sadler JE (1989) Structure of the gene for human von Willebrand factor. J Biol Chem 264:19514–19527
Marsden M, DeSimone DW (2001) Regulation of cell polarity, radial intercalation and epiboly in Xenopus: novel roles for integrin and fibronectin. Development 128:3635–3647
Massague J (1987) Identification of receptors for type-beta transforming growth factor. Methods Enzymol 146:174–195. doi:10.1016/S0076-6879(87)46020-5
McDonald NQ, Hendrickson WA (1993) A structural superfamily of growth factors containing a cystine knot motif. Cell 73:421–424. doi:10.1016/0092-8674(93)90127-C
Mercurio S, Latinkic B, Itasaki N, Krumlauf R, Smith JC (2004) Connective-tissue growth factor modulates WNT signalling and interacts with the WNT receptor complex. Development 131:2137–2147. doi:10.1242/dev.01045
Minamizato T, Sakamoto K, Liu T, Kokubo H, Katsube K, Perbal B, Nakamura S, Yamaguchi A (2007) CCN3/NOV inhibits BMP-2-induced osteoblast differentiation by interacting with BMP and Notch signaling pathways. Biochem Biophys Res Commun 354:567–573. doi:10.1016/j.bbrc.2007.01.029
Nakayama N, Han CE, Scully S, Nishinakamura R, He C, Zeni L, Yamane H, Chang D, Yu D, Yokota T, Wen D (2001) A novel chordin-like protein inhibitor for bone morphogenetic proteins expressed preferentially in mesenchymal cell lineages. Dev Biol 232:372–387. doi:10.1006/dbio.2001.0200
Notredame C, Higgins DG, Heringa J (2000) T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217. doi:10.1006/jmbi.2000.4042
O’Brien TP, Lau LF (1992) Expression of the growth factor-inducible immediate early gene cyr61 correlates with chondrogenesis during mouse embryonic development. Cell Growth Differ 3:645–654
O’Leary JM, Hamilton JM, Deane CM, Valeyev NV, Sandell LJ, Downing AK (2004) Solution structure and dynamics of a prototypical chordin-like cysteine-rich repeat (von Willebrand Factor type C module) from collagen IIA. J Biol Chem 279:53857–53866. doi:10.1074/jbc.M409225200
Paakkonen K, Tossavainen H, Permi P, Rakkolainen H, Rauvala H, Raulo E, Kilpelainen I, Guntert P (2006) Solution structures of the first and fourth TSR domains of F-spondin. Proteins 64:665–672. doi:10.1002/prot.21030
Perbal B (2001a) The CCN family of genes: a brief history. Mol Pathol 54:103–104. doi:10.1136/mp.54.2.103
Perbal B (2001b) NOV (nephroblastoma overexpressed) and the CCN family of genes: structural and functional issues. Mol Pathol 54:57–79. doi:10.1136/mp.54.2.57
Perbal B (2003) Communication is the key. Cell Commun Signal 1:3. doi:10.1186/1478-811X-1-3
Perbal B (2004) CCN proteins: multifunctional signalling regulators. Lancet 363:62–64. doi:10.1016/S0140-6736(03)15172-0
Perbal B (2006) The CCN3 protein and cancer. Adv Exp Med Biol 587:23–40
Perbal B, Planque N (2006) CCN3. In: Riley B (ed) AfCS-Nature Molecule Pages. http://www.signaling-gateway.org/
Perbal B, Takigawa M (2005) CCN proteins : a new family of cell growth and differentiation regulators 1st Edition. Imperial College Press
Perbal B, Martinerie C, Sainson R, Werner M, He B, Roizman B (1999) The C-terminal domain of the regulatory protein NOVH is sufficient to promote interaction with fibulin 1C: a clue for a role of NOVH in cell-adhesion signaling. Proc Natl Acad Sci USA 96:869–874. doi:10.1073/pnas.96.3.869
Planque N, Perbal B (2003) A structural approach to the role of CCN (CYR61/CTGF/NOV) proteins in tumourigenesis. Cancer Cell Int 3:15. doi:10.1186/1475-2867-3-15
Planque N, Long Li C, Saule S, Bleau AM, Perbal B (2006) Nuclear addressing provides a clue for the transforming activity of amino-truncated CCN3 proteins. J Cell Biochem 99:105–116. doi:10.1002/jcb.20887
Rider CC (2006) Heparin/heparan sulphate binding in the TGF-beta cytokine superfamily. Biochem Soc Trans 34:458–460. doi:10.1042/BST0340409
Roberts AB, Sporn MB, Assoian RK, Smith JM, Roche NS, Wakefield LM, Heine UI, Liotta LA, Falanga V, Kehrl JH et al (1986) Transforming growth factor type beta: rapid induction of fibrosis and angiogenesis in vivo and stimulation of collagen formation in vitro. Proc Natl Acad Sci USA 83:4167–4171. doi:10.1073/pnas.83.12.4167
Sakamoto K, Yamaguchi S, Ando R, Miyawaki A, Kabasawa Y, Takagi M, Li CL, Perbal B, Katsube K (2002) The nephroblastoma overexpressed gene (NOV/ccn3) protein associates with Notch1 extracellular domain and inhibits myoblast differentiation via Notch signaling pathway. J Biol Chem 277:29399–29405. doi:10.1074/jbc.M203727200
Sakuta H, Suzuki R, Takahashi H, Kato A, Shintani T, Iemura S, Yamamoto TS, Ueno N, Noda M (2001) Ventroptin: a BMP-4 antagonist expressed in a double-gradient pattern in the retina. Science 293:111–115. doi:10.1126/science.1058379
Schlunegger MP, Grutter MG (1993) Refined crystal structure of human transforming growth factor beta 2 at 1.95 A resolution. J Mol Biol 231:445–458. doi:10.1006/jmbi.1993.1293
Schuetze N, Wagemanns R, Schilling T, Jakob FJ (2006) CCN1. In: Riley B (ed) AfCS-Nature Molecule Pages. http://www.signaling-gateway.org/
Schultz-Cherry S, Chen H, Mosher DF, Misenheimer TM, Krutzsch HC, Roberts DD, Murphy-Ullrich JE (1995) Regulation of transforming growth factor-beta activation by discrete sequences of thrombospondin 1. J Biol Chem 270:7304–7310. doi:10.1074/jbc.270.13.7304
Sipes JM, Guo N, Negre E, Vogel T, Krutzsch HC, Roberts DD (1993) Inhibition of fibronectin binding and fibronectin-mediated cell adhesion to collagen by a peptide from the second type I repeat of thrombospondin. J Cell Biol 121:469–477. doi:10.1083/jcb.121.2.469
Sitar T, Popowicz GM, Siwanowicz I, Huber R, Holak TA (2006) Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins. Proc Natl Acad Sci USA 103:13028–13033. doi:10.1073/pnas.0605652103
Siwanowicz I, Popowicz GM, Wisniewska M, Huber R, Kuenkele KP, Lang K, Engh RA, Holak TA (2005) Structural basis for the regulation of insulin-like growth factors by IGF binding proteins. Structure 13:155–167. doi:10.1016/j.str.2004.11.009
Stndker L, Braulke T, Mostafavi H, Meyer M, Hning S, Gimnez-Gallego G, Forsmann W (2000) Partial IGF Affinity of Circulating N- and C-Terminal Fragments of Human Insulin-like Growth Factor Binding Protein-4 (IGFBP-4) and the Disulphide Bonding Pattern of the C-terminal IGFBP-4 Domain. Biochemistry 39:7
Subramaniam MM, Lazar N, Navarro S, Perbal B, Llombart-Bosch A (2008) Expression of CCN3 protein in human Wilms' tumors: immunohistochemical detection of CCN3 variants using domain-specific antibodies. Virchows Arch 452:33–39. doi:10.1007/s00428-007-0523-3
Takagi J, Fujisawa T, Usui T, Aoyama T, Saito Y (1993) A single chain 19-kDa fragment from bovine thrombospondin binds to type V collagen and heparin. J Biol Chem 268:15544–15549
Tan K, Duquette M, Liu JH, Dong Y, Zhang R, Joachimiak A, Lawler J, Wang JH (2002) Crystal structure of the TSP-1 type 1 repeats: a novel layered fold and its biological implication. J Cell Biol 159:373–382. doi:10.1083/jcb.200206062
Tanaka S, Sugimachi K, Saeki H, Kinoshita J, Ohga T, Shimada M, Maehara Y (2001) A novel variant of WISP1 lacking a Von Willebrand type C module overexpressed in scirrhous gastric carcinoma. Oncogene 20:5525–5532. doi:10.1038/sj.onc.1204723
Tanaka S, Sugimachi K, Shimada M, Maehara Y (2002) Variant WISPs as targets for gastrointestinal carcinomas. Gastroenterology 123:392–393. doi:10.1053/gast.2002.34589
Thorstensen L, Diep CB, Meling GI, Aagesen TH, Ahrens CH, Rognum TO, Lothe RA (2001) WNT1 inducible signaling pathway protein 3, WISP-3, a novel target gene in colorectal carcinomas with microsatellite instability. Gastroenterology 121:1275–1280. doi:10.1053/gast.2001.29570
Todorovicc V, Chen CC, Hay N, Lau LF (2005) The matrix protein CCN1 (CYR61) induces apoptosis in fibroblasts. J Cell Biol 171:559–568. doi:10.1083/jcb.200504015
Tong ZY, Brigstock DR (2006) Intrinsic biological activity of the thrombospondin structural homology repeat in connective tissue growth factor. J Endocrinol 188:R1–R8. doi:10.1677/joe.1.06719
Tossavainen H, Pihlajamaa T, Huttunen TK, Raulo E, Rauvala H, Permi P, Kilpelainen I (2006) The layered fold of the TSR domain of P. falciparum TRAP contains a heparin binding site. Protein Sci 15:1760–1768. doi:10.1110/ps.052068506
Tucker RP (2004) The thrombospondin type 1 repeat superfamily. Int J Biochem Cell Biol 36:969–974. doi:10.1016/j.biocel.2003.12.011
Voorberg J, Fontijn R, Calafat J, Janssen H, van Mourik JA, Pannekoek H (1991) Assembly and routing of von Willebrand factor variants: the requirements for disulfide-linked dimerization reside within the carboxy-terminal 151 amino acids. J Cell Biol 113:195–205. doi:10.1083/jcb.113.1.195
Wong M, Kireeva ML, Kolesnikova TV, Lau LF (1997) Cyr61, product of a growth factor-inducible immediate-early gene, regulates chondrogenesis in mouse limb bud mesenchymal cells. Dev Biol 192:492–508. doi:10.1006/dbio.1997.8766
Yamanaka Y, Wilson EM, Rosenfeld RG, Oh Y (1997) Inhibition of insulin receptor activation by insulin-like growth factor binding proteins. J Biol Chem 272:30729–30734. doi:10.1074/jbc.272.49.30729
Yan X, Baxter RC, Perbal B, Firth SM (2006) The aminoterminal insulin-like growth factor (IGF) binding domain of IGF binding protein-3 cannot be functionally substituted by the structurally homologous domain of CCN3. Endocrinology 147:5268–5274. doi:10.1210/en.2005-1568
Zhang Y, Pan Q, Zhong H, Merajver SD, Kleer CG (2005) Inhibition of CCN6 (WISP3) expression promotes neoplastic progression and enhances the effects of insulin-like growth factor-1 on breast epithelial cells. Breast Cancer Res 7:R1080–R1089. doi:10.1186/bcr1351
Zhang JL, Huang Y, Qiu LY, Nickel J, Sebald W (2007) von Willebrand factor type C domain-containing proteins regulate bone morphogenetic protein signaling through different recognition mechanisms. J Biol Chem 282:20002–20014. doi:10.1074/jbc.M700456200
Zhang JL, Qiu LY, Kotzsch A, Weidauer S, Patterson L, Hammerschmidt M, Sebald W, Mueller TD (2008) Crystal structure analysis reveals how the Chordin family member crossveinless 2 blocks BMP-2 receptor binding. Dev Cell 14:739–750. doi:10.1016/j.devcel.2008.02.017
Zhu Y, Oganesian A, Keene DR, Sandell LJ (1999) Type IIA procollagen containing the cysteine-rich amino propeptide is deposited in the extracellular matrix of prechondrogenic tissue and binds to TGF-beta1 and BMP-2. J Cell Biol 144:1069–1080. doi:10.1083/jcb.144.5.1069
Acknowledgements
The ‘Angiogenesis Research’ in KRA’s laboratory is supported by the Wellcome Trust (UK) through a programme grant (number 083191). KRA wishes to thank the Royal Society (UK) for an Industry Fellowship. The research in BP’s laboratory at the University Paris 7 D. Diderot was funded by a European contract ‘PROTHETS (Prognosis and therapeutic targets of Ewing's family of tumours) FP6 contract number 503036’ and the French ministry of Education.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Holbourn, K.P., Perbal, B. & Ravi Acharya, K. Proteins on the catwalk: modelling the structural domains of the CCN family of proteins. J. Cell Commun. Signal. 3, 25–41 (2009). https://doi.org/10.1007/s12079-009-0048-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12079-009-0048-4