Abstract
The recent outbreak of coronavirus disease (COVID-19) caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has already affected a large population of the world. SARS-CoV-2 belongs to the same family of severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV). COVID-19 has a complex pathology involving severe acute respiratory infection, hyper-immune response, and coagulopathy. At present, there is no therapeutic drug or vaccine approved for the disease. There is an urgent need for an ideal animal model that can reflect clinical symptoms and underlying etiopathogenesis similar to COVID-19 patients which can be further used for evaluation of underlying mechanisms, potential vaccines, and therapeutic strategies. The current review provides a paramount insight into the available animal models of SARS-CoV-2, SARS-CoV, and MERS-CoV for the management of the diseases.
Similar content being viewed by others
Introduction
In the past two decades, two outbreaks of coronaviruses (CoVs) in Asian subcontinent and the Middle East increased the chances of such outbreak in the near future. The severe acute respiratory syndrome coronavirus (SARS-CoV) outbreak at the end of 2002 (November) in China in which 8,098 confirmed cases were reported with 774 total deaths (9.6%) (Ksiazek et al.2003; WHO 2002). Another coronavirus i.e. Middle East respiratory syndrome (MERS) occurred in 2012 with a fatality rate of around 35% (WHO 2019; Bermingham et al.2012). Recently, a new coronavirus called as novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) outbreak in December, 2019, in China. There is an emergence of global health threats and the situation became worse at the global level (Rodriguez-Morales et al.2020). The World Health Organization (WHO) termed the syndrome caused by the novel coronavirus as “COVID-19” on 11th, Feb 2020 (Gralinski and Menachery 2020).
Coronaviruses fall in Coronaviridae family and subfamily Coronavirinae, the members of which infect a broad range of hosts, producing symptoms and diseases of wide spectrum from a flu like to severe acute respiratory infection (SARI). The SARS-CoV-2 is considered to be one of the seven members of the CoV family that infect humans (Zhu et al.2020). Similar to other coronaviruses, the transmission of SARS-CoV-2 among humans occurs through droplets, direct contact, any physical objects, and aerosols (WHO 2020a; Liu et al.2020). Owing to the rapid transmission, SARS-CoV-2 epidemic has become a global public health challenge. On March 11, COVID-19 outbreak was declared as a pandemic by WHO (Cucinotta and Vanelli 2020). By June 10, more than 7 million people have been globally affected, with nearly 408,025 fatalities (WHO 2020b).
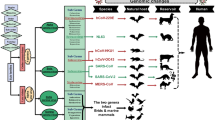
Due to the high morbidity and mortality rate of coronaviruses infection, there is an unmet need for therapeutic strategies to treat these diseases (Sarma et al.2020). Currently, there are no recommended therapeutic drugs or vaccine for coronaviruses. This emphasizes the surge for a suitable animal model to explore the pathogenesis and evaluation of countermeasures for the disease. With the outbreak of the coronaviruses, numerous animal models were developed in the past (Gralinski and Baric 2015). The current review highlighted the animal models of coronaviruses, summarized the strength and weaknesses of the existing animal models along with their key features.
Animal Models for SARS-CoV-2
Non-Human Primate Models
SARS-CoV-2 was recently recognized as a novel coronavirus causing the symptoms from mild to critical type (SARI). Munster et al. used SARS-CoV-2 infected rhesus macaques to study the pathogenesis of COVID-19. They observed high viral titers in nose swabs, throat swabs, and also lung lesions at varying degrees in all animals (Table 1). This model recapitulates COVID-19 symptoms and can be used further to elucidate the therapy for SARS-CoV-2 infection (Munster et al.2020).
Rockx et al. compared the pathogenesis of SARS and MERS with COVID-19 by inoculating cynomolgus macaques with virus infection (Rockx et al.2020). The virus samples were taken from the throat and nose. The diffuse alveolar damage was reported. The results of the study thus demonstrated that the infection with SARS-CoV-2 in non-human primates induced signs that resemble COVID-19 like disease, and the severity of virus infection lies in between the MERS and SARS (Mahase 2020).
Yu et al. studied the association of virus infection with age. The viral strain was inoculated in rhesus macaques of 3–5 years old and 15 years old through intra-tracheal route. The clinical signs, for instance, viral replication, and histopathological changes were analyzed. Replication of virus was more in lungs and nasopharyngeal swabs of old rhesus macaque as compared to young rhesus macaque after infection. Old rhesus macaques also observed to have diffuse severe interstitial pneumonia (Yu et al.2020).
In a comparative study, three species of non-human primates were infected with SARS-CoV-2 (rhesus macaque, Macaca fascicularis, and common marmoset). Severe gross lesions and histopathological changes in the vital organs of all animals were observed. The study found that the rhesus macaque model was more susceptible to SARS-CoV-2, as compared to Macaca fascicularis and common marmoset (Lu et al.2020).
Woolsey et al. determined that African green monkeys supported a high level of SARS-CoV-2 replication and developed a respiratory tract related illnesses that might be more substantial than reported for other non-human primate species like cynomolgus and rhesus macaques. The study also reported high viral loads in feces and in mucosal samples of all monkeys after 15 days post-infection (dpi) (Woolsey et al.2020) (Table 1).
Mouse Models
The inoculation of BALB/c mice with mouse-adapted SARS-CoV-2 at passage 6 (MACSp6) significantly affected all groups of mice irrespective of their ages. The infection resulted in moderate pneumonia and inflammation. A study reported higher viral load in lungs along with significant histopathological changes like denatured trachea, inflammation in pulmonary alveoli, detection of viral antigen in the trachea, bronchiole, in type II pneumocytes. There was a significant elevation of inflammatory chemokine and cytokines in sera and lung macrophages similar to clinical symptoms (Gu et al.2020).
Dinnon et al. constructed a recombinant virus (SARS-CoV-2 MA) that could replicate in both upper as well as lower airways of all groups of BALB/c mice irrespective of their ages. The results showed that there was more severe infection in aged mice in comparison to young mice (Dinnon et al.2020).
A study has been done based on the human ACE2 (hACE2) transgenic mice to investigate the pathogenesis of COVID-19. Infected mice reported weight loss and increased viral titer in lungs, and histopathological changes revealed the presence of interstitial pneumonia. The mice model seems to be promising for the evaluation of therapeutic measures for COVID-19 (Bao et al.2020).
One of the studies examined the infectivity and pathological changes in SARS-CoV-2 infected transgenic hACE2 mice, which was developed using lung ciliated epithelial cell-specific HFH4/FOXJ1 promoter 81. The model showed a partial simulation of the human COVID-19. The significant decrease in body weight, interstitial pneumonia, and fatality along with the involvement of other organs like heart, brain, and eye depicted its similarity to the human counterpart. The study also showed protection on reinfection in the survived mice (Jiang et al.2020) (Table 1).
Sun SH et al. developed the hACE2 mouse model using CRISPR/Cas9 knock in technology to study the SARS-CoV-2 infection and compared it with the wild type C57BL/6 mice model. There was high viral titer in brain, lungs and trachea of hACE2 mice than wild type mice. SARS-CoV-2 infection in hACE2 mice associated with increased cytokine levels and interstitial pneumonia with no mortality. Moreover, intragastric inoculation of the SARS-CoV-2 infection altered significant pathological changes in lungs when compared to wild type mice (Sun et al.2020b).
Sun J et al. developed a transgenic mice model by delivery of human ACE2 exogenously with a replication deficient adenovirus (Ad5-hACE2). The inoculation of mice with SARS-CoV-2 infection linked to high viral load in lungs along with weight loss and pneumonia. The role of type-1 interferon, signal transducer and activator of transcription 1 (STAT1) signaling is vital in viral clearance of virus and infection. Further, the study concluded the beneficial role of using human convalescent plasma and two antiviral agents (polyinosinic: polycytidylic acid and Remdesivir) (Sun J et al.2020a).
Hassan et al. developed hACE2 transduced mice and infected these mice with SARS-CoV-2. The study observed significant weight loss, high viral load in lungs and severe lung pathology in infected mice (Hassan et al.2020).
Hamster Model
Chan et al. observed significant binding of the novel coronavirus spike with the ACE2 receptor of the Syrian Hamster by in silico study (Table 1). The viral load significantly increased in the hamster which leads to diffuse alveolar damage in the initial stage and extensive apoptosis in the later phase of infection (Chan et al.2020).
The clinical and histopathological observations from SARS-CoV-2 hamster model closely resemble to the clinical condition. The airway involvement is evident from nasal turbinate to the trachea and pulmonary alveoli, associated with changes of inflammation, cellular viral N protein expression, and high viral load during the first week. The disease progressed with increasing respiratory rate, decreasing activity, and progressive weight loss, and was found to be most severe by 6 dpi, which is similar to the disease course of COVID-19 patients (Huang et al.2020).
Sia et al. evaluated the transmission and pathogenesis of SARS-CoV-2 infection in the golden Syrian hamster model. The animal groups infected with SARS-CoV-2 showed significant weight loss, and viral replication in the respiratory tract. Viral load was high on 2 dpi and rapid clearance was observed on 7 dpi, which could be attributed to the presence of CD3 positive T-lymphocytes. The involvement of olfactory sensory neurons simulates the anosmia in COVID-19 cases. The transmission of SARS-CoV-2 to naïve hamsters by aerosols can be helpful for further studies (Sia et al.2020) (Table 1).
Ferrets Model
Kim et al. used ferrets to study the SARS-CoV-2 infection and its transmission. The study reported that the ferrets were more susceptible to infection and transmission of the virus from one ferret to naive ferrets by direct or indirect physical contact. They showed that infected ferrets exhibited more replication of the virus in the upper respiratory tract and it recapitulates COVID-19 human conditions and can be further used to evaluate the countermeasures for the disease (Kim et al.2020) (Table 1).
Animal Models for SARS-CoV
Non-Human Primate Models
For the development of SARS-CoV infection model, many species, like old and new world primates, were used. There was significant infection observed in cynomolgus (Fouchier et al.2003) and rhesus macaques (Rowe et al.2004), common marmosets (Greenough et al.2005) and African green monkeys (McAuliffe et al.2004), when the virus was inoculated within respiratory tract in those primates. However, in the case of squirrel monkeys and mustached tamarins, investigators were not able to induce infection (Roberts et al.2008; Subbarao and Roberts 2006).
There was significantly high viral replication in cynomolgus, rhesus, and African green monkeys (McAuliffe et al.2004). While in marmosets, diarrhea, pneumonitis, fever, watery stool, and hepatitis were observed (Greenough et al.2005). Investigators have variable findings as a number of factors may affect the results, including age and animal source, dose, route of administration of infection and inoculation of the virus, and their history of the environment (Subbarao and Roberts 2006).
However, results in out-bred species are not consistent because of biological variability. Therefore, it is important to carry large sample sizes for the study to find out the meaningful conclusions (Table 2).
Mouse Models
There were different mice models used for the study of infection. When BALB/c mice were infected intranasally, there was an increase in viral titer in the lungs but no sign of morbidity or mortality was noticed. After 2 to 3 dpi, virus replication significantly increased in the respiratory tract but started decreasing at day 5 dpi (Subbarao et al.2004). This showed that young BALB/c mice failed to exhibit clinical signs of disease but permitted viral replication. On the other hand, a study showed that one-year-old BALB/c mice exhibited critical infection as compared to younger mice after SARS-CoV infection (Vogel et al.2007). Histopathological findings such as bronchiolitis, diffuse alveolar damage, and patchy interstitial pneumonitis were observed in BALB/c mice model of SARS-CoV. It was also observed that old age BALB/C mice were more prone to this disease as similar to the clinical symptoms observed in elderly people in the 2003 SARS outbreak.
One of the studies suggested that 129S mice infected with SARS-CoV exhibited virus replication and self-limited bronchiolitis. 129S mice strain also reported mild weight loss and pneumonitis after SARS-CoV infection. While on the other side, STAT 1 -/- mice in the 129S background observed sustained replication of the virus and histopathological changes, which were mimicking the human’s disease. However, vaccine studies are limited where targeted mice have immune defects (Hogan et al.2004; Frieman et al.2010).
Moreover, C57BL/6 mice were also used as a SARS-CoV model. Virus isolates administered intranasal into C57BL/6 mice showed high replication in the respiratory tract of mice with a peak on the 3rd day and clearance on the 9 dpi. The study observed the presence of transient systemic infection in the lungs which ultimately affects the brain (Glass et al.2004) (Table 2).
Hamster Model
Another model used to study the SARS-CoV infection is a golden Syrian hamster. There was a significantly high titer of viruses in the lungs along with interstitial pneumonitis (Roberts et al.2005). Because of the clinical signs like pulmonary histopathology and high virus load, the hamster can be used as a model for the study of therapeutics, immunotherapy, and immunoprophylaxis. It was observed that viruses got cleared off by 7dpi. (Table 2).
Ferrets and Domestic Cat Model
Various studies showed that domestic cats and ferrets are prone to SARS-CoV infection because virus replication, as well as specific antibodies, were found in both the species. After indicating drowsiness and epiphysitis 16 to 21 dpi, ferrets died (Martina et al.2003). However, with domestic cats, no further study has been conducted. After the inoculation of infection via intranasal route, the replication of the virus in the lungs and histological changes like pneumonitis were reported (ter Meulen et al.2004) (Table 2).
Animal Models of MERS-CoV
There is a difference in the homology of human and small animals DPP4 molecules which is the main virus receptor of MERS-CoV. Therefore, mice, guinea pig, and ferrets are not usually permissive to the MERS-CoV (Cockrell et al.2014; Coleman et al.2014; de Wit et al.2013a, b; Raj et al.2013). However, non-human primates, rabbits, pigs, and dromedary camels are naturally permissive for the virus.
Dromedary Camel Model
Adney et al. studied the pathology and transmission of MERS-CoV among dromedary camels. The animals displayed slight increases in body temperature and rhinorrhea. A large quantity of nasal shedding of the virus is suggestive of an animal-to-animal and animal-to-human transmission which may occur through droplets or direct contact. MERS-CoV mainly affects the upper respiratory tract and mimics mild clinical signs. Since in clinical settings, the disease shows severe pathology, so camel as a model to study therapeutics is not suitable (Adney et al.2014) (Table 3).
Alpaca Model
MERS-CoV is classified under Biosafety Level 3 which poses difficulty in using camels for infection study. Alpaca, belonging to the Camelidae family, provides an alternative to camels. Crameri et al. used alpaca as a prospective substitute to a camel for MERS-CoV vaccine testing (Crameri et al.2016). On exposure to MERS-CoV, the study observed dissimilarity between the immune response of two species. However, the pathogenesis of infection in alpaca is unknown (Table 3).
Non-Human Primate Models
de Wit et al. used rhesus macaques to develop the MERS model. There was high viral load of MERS-CoV in the lower respiratory tract. The viral strain mainly affected type I and II pneumocytes that formed the architecture of the alveolar space. Since high expression of MERS-CoV was present in the lungs, therefore, there was limited virus shedding. There was no viral replication in the kidney. The study concluded that renal failure in humans might be associated with hypoxia but not with viral dissemination. Due to its transitory nature, this model of rhesus macaques was not found to be exactly mimicking the critical clinical cases (de Wit et al.2013a). Similarly, Yao et al. observed a mild increase in temperature in rhesus monkeys. There was no virus detected in the upper respiratory tract. Significant pneumonia was observed with no systemic dissemination of the virus. The study concluded only lower respiratory infection with no extra-pulmonary effect (Yao et al.2014).
Human exposure to MERS-CoV leads to severe respiratory disease which might result in fatality and it is not easy to attain the same fatality rate in non-human primate models. Cockrell et al. used an infectious clone of novel MERS-CoV (icMERS-0) to induce MERS in the rhesus macaque. The clone strain was found to have higher pathogenicity in comparison to its wild type. MERS-CoV peaked at 3 to 5 dpi and resolved to 30 dpi. The respiratory disease was found to be placid and brief which resolves by day 30 dpi. The model mimics only mild infection of MERS in humans (Cockrell et al.2018).
In most of the cases, fatality is associated with comorbidities in MERS affected patients (Hui et al.2014; Saad et al.2014) and severity is linked to the immunocompromised state. In order to study the immune response and disease severity, Prescott et al. used the immunosuppressive drugs to down-regulate the immunity of rhesus macaques. The immunosuppressive state was found to be associated with a high titer of MERS-CoV but not supported by the pathology. In lung tissues, inflammatory cells significantly decreased in immunocompromised animals when evaluated histopathologically. This, in turn, shows the effect of the immune status of an individual on the shedding of the viral load. This study depicted the importance of considering the immune response along with using therapy to control infection in clinical settings (Prescott et al.2018) (Table 3).
Marmosets are more prone to MERS-CoV infection than the rhesus macaques and it might be suitable to screen therapeutic strategies for the disease in marmoset (Johnson et al.2015; Falzarano et al.2014). The study by Falzarano et al. displayed an increase in severity following exposure to MERS-CoV in marmoset than rhesus macaque. The titer was almost 1000 times greater in marmoset than in rhesus macaque. The infection persisted for a longer duration as compared in rhesus macaque and need euthanasia. In fact, there was a systemic dissemination of the virus. But there was no viral dissemination in the kidney. The study observed that the marmoset model recapitulates more closely to human MERS (Falzarano et al.2014).
Johnson et al. used two clones of MERS-CoV i.e. MERS-CoV-Jordan-n3/2012 and MERS-CoV-EMC/2012. The inoculation of MERS-CoV into common marmosets resulted in mild to moderate clinical presentation of the disease which might be due to the manipulations of the marmoset to a certain extent (Johnson et al.2015) (Table 3).
Yorkshire Landrace Pig Model
Pigs are naturally permissive to the MERS-CoV infection (de Wit et al.2017). Vergara-Alert et al. used HCoVEMC/2012 to infect Yorkshire Landrace pigs. The study observed no animal-to-animal transmission though RNA dissemination of the virus was significantly high. Thus, this model cannot be used to study therapeutic strategies as it does not depict the true clinical condition (Vergara-Alert et al.2017) (Table 3).
New Zealand White Rabbit Model
Houser et al. studied the effect of re-infection in the rabbits. The model supported the replication of MERS-CoV-EMC/2012 virus. The model displayed asymptomatic infection which is not associated with any significant clinical sign. The model is not suitable for preclinical screening of therapeutics or vaccines for MERS (Houser et al.2017) (Table 3).
Mouse Models
Mice are easily available, cost-effective, easy to handle, and suitable for initial screening of drug therapies and vaccines. Since, mice are not susceptible to MERS-CoV infection (de Wit et al.2013b; Coleman et al.2014), therefore Zhao et al. developed a mouse model for MERS using a recombinant adenovirus that expresses humanized dipeptidyl peptidase-4 (hDPP4) receptor (Zhao et al.2014). The study concluded significant weight loss and virus dissemination in the respiratory tract. Further interstitial pneumonia was observed in the mice. The transient hDPP4 expression, mild respiratory system involvement with no fatality are the drawbacks of the model (Zhao et al.2014).
Though the previous study used an adenovirus vector to develop the DPP4 receptor in mice vulnerable to MERS-CoV infection, it did not mimic more closely to the clinical conditions. Agrawal et al. used tissue-specific and inducible promoters, to derive humanized DPP4 transgenic mice. On exposure to MERS-CoV, the model showed high susceptibility to the virus (Agrawal et al.2015). Briefly, the transgenic model depicted progressive pneumonia with fatality.
Agrawal et al. developed a hCD26/DPP4 transgenic mice and the receptors were distributed throughout the body which was not physiologically relevant (Agrawal et al.2015). Pascal et al. used Veloci Gene technology to rapidly develop a new humanized model. The model showed active viral replication in the respiratory tract without cerebral infection (Pascal et al.2015). Tao et al. observed constant inflammatory infiltrates in both brain and lungs in humanized CD26 (hCD26)/DPP4 mice. The high viral titer was observed in the lungs and in the brain of mice on exposure to MERS-CoV infection. The pathological examination found gross and microscopic inflammation which was not restricted to lungs but a sign of systemic dissemination along with fatality in 4–6 dpi (Tao et al.2015).
Tao et al. showed acute severe infection which hampers the opportunity to fully understand the pathogenesis. Zhao et al. developed a mouse model that exhibits the codon-optimized- hDPP4 receptor which on exposure to viral strain showed severe disease pathology in lungs which further affected the brain and kidney. The severe infection and aberrant immune response lead to mortality. The exact mechanism of the severity of the disease model requires further evaluation (Zhao et al.2015).
Cockrell et al. used CRISPR–Cas9 technology to genetically alter a non-permissive host receptor. The model showed severe respiratory distress and fatality response to MERS-CoV. This was further supported by plethysmography that showed decreased pulmonary function which might be due to the inflammation in pneumocytes and airway epithelial cells. The viral titers were not detected in the brain thereby nullifying any confounding factors which could lead to fatality (Cockrell et al.2016).
Li et al. developed transgenic mice, and inoculated with mouse adapted MERS-CoV (MERSMA) viral strain which caused weight loss, infection in airway epithelia, pneumocytes, and macrophages. Though the model was rapid to develop, required no prior sensitization, the pathological changes were exclusively associated with the species-adapted mutations of the MERSMA clone. This hinders the model to fully mimic clinical conditions (Li et al.2017).
Fan et al. used CRISPR/Cas9 gene-editing technology to develop a knock-in-model of mice. The study displayed a high level of expression of hDPP4 in the respiratory tract and to a lesser extent in the brain. There is a clear site of infection along with acute respiratory distress syndrome (ARDS) in the lungs. There is also the dissemination of viruses into the central nervous system (CNS). The study observed high repeatability and the safety of the model (Fan et al.2018).
Iwata-Yoshikawa et al. developed a transgenic mouse model using the endogenous promoter. The hDPP4 mice model showed a high viral titer in the lower respiratory tract. Acute multifocal interstitial pneumonia was observed at 7 dpi. The viral titer was also observed in peripheral blood and lymphoid tissues. There was an age-associated immune response to exposure to the viral strain. The model expressed mild infection pathology. Moreover, after the infection, this model had not shown any signs of lesions in the brain and kidney (Iwata-Yoshikawa et al.2019) (Table 3).
Conclusions
-
1.
The coronaviruses pathology is associated with the involvement of severe acute respiratory infection (SARI) and immune deregulation, thus, it is requisite to perform a study using an assay system that must involve all the cell signalling and thus, using animal model is an unparallel approach.
-
2.
There are some animal models that recapitulate mild to moderate pathology, however, the clinical condition is related to high mortality and morbidity. Therefore, animal models which mimic critical pathologies are much closer to clinical conditions.
-
3.
The novel coronavirus (COVID-19) pathology is linked to viral respiratory infection, hyper-immune response, and coagulopathy (Lin et al.2020; Connors and Levy 2020), therefore, to understand the mechanism or to evaluate therapeutic countermeasures, the animal models should involve all these interplays in a single model. Since with the advent of technology, transgenic mice are the significant tool but the transgenic mice lack the coagulopathy component which is indispensable in order to understand the complete mechanism and to evaluate the countermeasures.
-
4.
Further, validation of the animal model is crucial. The error in the animal experimental study narrows the chances of the potential drugs or repurposing or repositioning drugs or vaccines to translate successfully to clinics and moreover, it is a wastage of resources. Thus, it is the need of the hour to validate the animal model using different criteria, for instance, face, construct, and predictive validity (Denayer et al.2014).
References
Adney DR, van Doremalen N, Brown VR, Bushmaker T, Scott D, de Wit E, Bowen RA, Munster VJ (2014) Replication and shedding of MERS-CoV in upper respiratory tract of inoculated dromedary camels. Emerg Infect Dis 20:1999–2005
Agrawal AS, Garron T, Tao X, Peng BH, Wakamiya M, Chan TS, Couch RB, Tseng CT (2015) Generation of a transgenic mouse model of Middle East respiratory syndrome coronavirus infection and disease. J Virol 89:3659–3670
Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, Wei Q, Yu P, Xu Y, Qi F, Qu Y, Li F, Lv Q, Wang W, Xue J, Gong S, Liu M, Wang G, Wang S, Song Z, Zhao L, Liu P, Zhao L, Ye F, Wang H, Zhou W, Zhu N, Zhen W, Yu H, Zhang X, Guo L, Chen L, Wang C, Wang Y, Wang X, Xiao Y, Sun Q, Liu H, Zhu F, Ma C, Yan L, Yang M, Han J, Xu W, Tan W, Peng X, Jin Q, Wu G, Qin C (2020) The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. https://doi.org/10.1038/s41586-020-2312-y
Bermingham A, Chand MA, Brown CS, Aarons E, Tong C, Langrish C, Hoschler K, Brown K, Galiano M, Myers R, Pebody RG, Green HK, Boddington NL, Gopal R, Price N, Newsholme W, Drosten C, Fouchier RA, Zambon M (2012) Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill 17:20290
Chan JF, Zhang AJ, Yuan S, Poon VK, Chan CC, Lee AC, Chan WM, Fan Z, Tsoi HW, Wen L, Liang R, Cao J, Chen Y, Tang K, Luo C, Cai JP, Kok KH, Chu H, Chan KH, Sridhar S, Chen Z, Chen H, To KK, Yuen KY (2020) Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. https://doi.org/10.1093/cid/ciaa325
Cockrell AS, Johnson JC, Moore IN, Liu DX, Bock KW, Douglas MG, Graham RL, Solomon J, Torzewski L, Bartos C, Hart R, Baric RS, Johnson RF (2018) A spike-modified Middle East respiratory syndrome coronavirus (MERS-CoV) infectious clone elicits mild respiratory disease in infected rhesus macaques. Sci Rep 8:10727
Cockrell AS, Peck KM, Yount BL, Agnihothram SS, Scobey T, Curnes NR, Baric RS, Heise MT (2014) Mouse dipeptidyl peptidase 4 is not a functional receptor for Middle East respiratory syndrome coronavirus infection. J Virol 88:5195–5199
Cockrell AS, Yount BL, Scobey T, Jensen K, Douglas M, Beall A, Tang XC, Marasco WA, Heise MT, Baric RS (2016) A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat Microbiol 2:16226
Coleman CM, Matthews KL, Goicochea L, Frieman MB (2014) Wild-type and innate immune-deficient mice are not susceptible to the Middle East respiratory syndrome coronavirus. J Gen Virol 95:408–412
Connors JM, Levy JH (2020) COVID-19 and its implications for thrombosis and anticoagulation. Blood 135:2033–2040
Crameri G, Durr PA, Klein R, Foord A, Yu M, Riddell S, Haining J, Johnson D, Hemida MG, Barr J, Peiris M, Middleton D, Wang LF (2016) Experimental infection and response to rechallenge of alpacas with middle east respiratory syndrome coronavirus. Emerg Infect Dis 22:1071–1074
Cucinotta D, Vanelli M (2020) WHO declares COVID-19 a pandemic. Acta Biomed 91:157–160
de Wit E, Prescott J, Baseler L, Bushmaker T, Thomas T, Lackemeyer MG, Martellaro C, Milne-Price S, Haddock E, Haagmans BL, Feldmann H, Munster VJ (2013a) The Middle East respiratory syndrome coronavirus (MERS-CoV) does not replicate in Syrian hamsters. PLoS ONE 8:e69127
de Wit E, Rasmussen AL, Falzarano D, Bushmaker T, Feldmann F, Brining DL, Fischer ER, Martellaro C, Okumura A, Chang J, Scott D, Benecke AG, Katze MG, Feldmann H, Munster VJ (2013b) Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques. Proc Natl Acad Sci U S A 110:16598–16603
de Wit E, Feldmann F, Horne E, Martellaro C, Haddock E, Bushmaker T, Rosenke K, Okumura A, Rosenke R, Saturday G, Scott D, Feldmann H (2017) Domestic Pig Unlikely Reservoir for MERS-CoV. Emerg Infect Dis 23(6):985–988
Denayer T, Stöhr T, Van Roy M (2014) Animal models in translational medicine: validation and prediction. New Horiz Transl Med 2:5–11
Dinnon KH, Leist SR, Schäfer A, Edwards CE, Martinez DR, Montgomery SA, West A, Yount BL, Hou YJ, Adams LE, Gully KL, Brown AJ, Huang E, Bryant MD, Choong IC, Glenn JS, Gralinski LE, Sheahan TP, Baric RS (2020) A mouse-adapted SARS-CoV-2 model for the evaluation of COVID-19 medical countermeasures. bioRxiv. https://doi.org/10.1101/2020.05.06.081497
Falzarano D, de Wit E, Feldmann F, Rasmussen AL, Okumura A, Peng X, Thomas MJ, van Doremalen N, Haddock E, Nagy L, LaCasse R, Liu T, Zhu J, McLellan JS, Scott DP, Katze MG, Feldmann H, Munster VJ (2014) Infection with MERS-CoV causes lethal pneumonia in the common marmoset. PLoS Pathog 10:e1004250
Fan C, Wu X, Liu Q, Li Q, Liu S, Lu J, Yang Y, Cao Y, Huang W, Liang C, Ying T, Jiang S, Wang Y (2018) A Human DPP4-Knockin Mouse's Susceptibility to Infection by Authentic and Pseudotyped MERS-CoV. Viruses 10:448
Fouchier RA, Kuiken T, Schutten M, van Amerongen G, van Doornum GJ, van den Hoogen BG, Peiris M, Lim W, Stöhr K, Osterhaus AD (2003) Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423:240
Frieman MB, Chen J, Morrison TE, Whitmore A, Funkhouser W, Ward JM, Lamirande EW, Roberts A, Heise M, Subbarao K, Baric RS (2010) SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog 6:e1000849
Glass WG, Subbarao K, Murphy B, Murphy PM (2004) Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J Immunol 173:4030–4039
Gralinski LE, Baric RS (2015) Molecular pathology of emerging coronavirus infections. J Pathol 235:185–195
Gralinski LE, Menachery VD (2020) Return of the Coronavirus: 2019-nCoV. Viruses 12:135
Greenough TC, Carville A, Coderre J, Somasundaran M, Sullivan JL, Luzuriaga K, Mansfield K (2005) Pneumonitis and multi-organ system disease in common marmosets (Callithrixjacchus) infected with the severe acute respiratory syndrome-associated coronavirus. Am J Pathol 167:455–463
Gu H, Chen Q, Yang G, He L, Fan H, Deng YQ, Wang Y, Teng Y, Zhao Z, Cui Y, Li Y, Li XF, Li J, Zhang N, Yang X, Chen S, Zhao G, Wang X, Luo D, Wang H, Yang X, Li Y, Han G, He Y, Zhou X, Geng S, Sheng X, Jiang S, Sun S, Qin, CF, Zhou Y (2020) Rapid adaptation of SARS-CoV-2 in BALB/c mice: Novel mouse model for vaccine efficacy. bioRxiv.https://doi.org/10.1101/2020.05.02.073411
Hassan AO, Case JB, Winkler ES, Thackray LB, Kafai NM, Bailey AL, McCune BT, Fox JM, Chen RE, Alsoussi WB, Turner JS, Schmitz AJ, Lei T, Shrihari S, Keeler SP, Fremont DH, Greco S, McCray PB, Perlman S, Holtzman MJ, Ellebedy AH, Diamond MS (2020) A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell. https://doi.org/10.1016/j.cell.2020.06.011
Hogan RJ, Gao G, Rowe T, Bell P, Flieder D, Paragas J, Kobinger GP, Wivel NA, Crystal RG, Boyer J, Feldmann H, Voss TG, Wilson JM (2004) Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J Virol 78:11416–11421
Houser KV, Broadbent AJ, Gretebeck L, Vogel L, Lamirande EW, Sutton T, Bock KW, Minai M, Orandle M, Moore IN, Subbarao K (2017) Enhanced inflammation in New Zealand white rabbits when MERS-CoV reinfection occurs in the absence of neutralizing antibody. PLoS Pathog 13:e1006565
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506
Hui DS, Memish ZA, Zumla A (2014) Severe acute respiratory syndrome vs. the Middle East respiratory syndrome. Curr Opin Pulm Med 20:233–241
Iwata-Yoshikawa N, Okamura T, Shimizu Y, Kotani O, Sato H, Sekimukai H, Fukushi S, Suzuki T, Sato Y, Takeda M, Tashiro M, Hasegawa H, Nagata N (2019) Acute respiratory infection in human dipeptidyl peptidase 4-transgenic mice infected with middle east respiratory syndrome coronavirus. J Virol 93:e01818
Jiang RD, Liu MQ, Chen Y, Shan C, Zhou YW, Shen XR, Li Q, Zhang L, Zhu Y, Si HR, Wang Q, Min J, Wang X, Zhang W, Li B, Zhang HJ, Baric RS, Zhou P, Yang XL, Shi ZL (2020) Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. https://doi.org/10.1016/j.cell.2020.05.027
Johnson RF, Via LE, Kumar MR, Cornish JP, Yellayi S, Huzella L, Postnikova E, Oberlander N, Bartos C, Ork BL, Mazur S, Allan C, Holbrook MR, Solomon J, Johnson JC, Pickel J, Hensley LE, Jahrling PB (2015) Intratracheal exposure of common marmosets to MERS-CoV Jordan-n3/2012 or MERS-CoV EMC/2012 isolates does not result in lethal disease. Virology 485:422–430
Kim YI, Kim SG, Kim SM, Kim EH, Park SJ, Yu KM, Chang JH, Kim EJ, Lee S, Casel MAB, Um J, Song MS, Jeong HW, Lai VD, Kim Y, Chin BS, Park JS, Chung KH, Foo SS, Poo H, Mo IP, Lee OJ, Webby RJ, Jung JU, Choi YK (2020) Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microb 27:704–709.e2
Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group (2003) A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348:1953–1966
Kuiken T, Fouchier RA, Schutten M, Rimmelzwaan GF, van Amerongen G, van Riel D, Laman JD, de Jong T, van Doornum G, Lim W, Ling AE, Chan PK, Tam JS, Zambon MC, Gopal R, Drosten C, van der Werf S, Escriou N, Manuguerra JC, Stöhr K, Peiris JS, Osterhaus AD (2003) Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263–270
Li K, Wohlford-Lenane CL, Channappanavar R, Park JE, Earnest JT, Bair TB, Bates AM, Brogden KA, Flaherty HA, Gallagher T, Meyerholz DK, Perlman S, McCray PB Jr (2017) Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc Natl Acad Sci U S A 114:E3119–E3128
Lin L, Lu L, Cao W, Li T (2020) Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg Microb Infect 9:727–732
Liu Y, Ning Z, Chen Y, Guo M, Liu Y, Gali NK, Sun L, Duan Y, Cai J, Westerdahl D, Liu X, Xu K, Ho KF, Kan H, Fu Q, Lan K (2020) Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. https://doi.org/10.1038/s41586-020-2271-3
Lu S, Zhao Y, Yu W, Yang Y, Gao J, Wang J, Kuang D, Yang M, Yang J, Ma C, Xu J, Qian, X, Li H, Zhao S, Li J, Wang H, Long H, Zhou J, Luo F, Ding K, Wu D, Zhang Y, Dong Y, Liu Y, Zheng Y, Lin X, Jiao L, Zheng H, Dai Q, Sun Q, Hu Y, Ke C, Liu H, Peng X (2020) Comparison of SARS-CoV-2 infections among 3 species of non-human primates. bioRxiv. https://doi.org/10.1101/2020.04.08.031807
Mahase E (2020) Coronavirus covid-19 has killed more people than SARS and MERS combined, despite lower case fatality rate. BMJ 368:m641
Martina BE, Haagmans BL, Kuiken T, Fouchier RA, Rimmelzwaan GF, Van Amerongen G, Peiris JS, Lim W, Osterhaus AD (2003) Virology: SARS virus infection of cats and ferrets. Nature 425:915
McAuliffe J, Vogel L, Roberts A, Fahle G, Fischer S, Shieh WJ, Butler E, Zaki S, St Claire M, Murphy B, Subbarao K (2004) Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology 330:8–15
Munster VJ, Feldmann F, Williamson BN, van Doremalen N, Pérez-Pérez L, Schulz J, Meade-White K, Okumura A, Callison J, Brumbaugh B, Avanzato VA, Rosenke R, Hanley PW, Saturday G, Scott D, Fischer ER, de Wit E (2020) Respiratory disease and virus shedding in rhesus macaques inoculated with SARS-CoV-2. bioRxiv preprint.https://doi.org/10.1101/2020.03.21.001628
Pascal KE, Coleman CM, Mujica AO, Kamat V, Badithe A, Fairhurst J, Hunt C, Strein J, Berrebi A, Sisk JM, Matthews KL, Babb R, Chen G, Lai KM, Huang TT, Olson W, Yancopoulos GD, Stahl N, Frieman MB, Kyratsous CA (2015) Pre- and post-exposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc Natl Acad Sci U S A 112:8738–8743
Prescott J, Falzarano D, de Wit E, Hardcastle K, Feldmann F, Haddock E, Scott D, Feldmann H, Munster VJ (2018) Pathogenicity and viral shedding of MERS-CoV in immunocompromised rhesus macaques. Front Immunol 9:205
Qin C, Wang J, Wei Q, She M, Marasco WA, Jiang H, Tu X, Zhu H, Ren L, Gao H, Guo L, Huang L, Yang R, Cong Z, Guo L, Wang Y, Liu Y, Sun Y, Duan S, Qu J, Chen L, Tong W, Ruan L, Liu P, Zhang H, Zhang J, Zhang H, Liu D, Liu Q, Hong T, He W (2005) An animal model of SARS produced by infection of Macacamulatta with SARS coronavirus. J Pathol 206:251–259
Raj VS, Mou H, Smits SL, Dekkers DH, Müller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL (2013) Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254
Roberts A, Lamirande EW, Vogel L, Jackson JP, Paddock CD, Guarner J, Zaki SR, Sheahan T, Baric R, Subbarao K (2008) Animal models and vaccines for SARS-CoV infection. Virus Res 133:20–32
Roberts A, Vogel L, Guarner J, Hayes N, Murphy B, Zaki S, Subbarao K (2005) Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J Virol 79:503–511
Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, Oude Munnink BB, de Meulder D, van Amerongen G, van den Brand J, Okba NMA, Schipper D, van Run P, Leijten L, Sikkema R, Verschoor E, Verstrepen B, Bogers W, Langermans J, Drosten C, Fentener van Vlissingen M, Fouchier R, de Swart R, Koopmans M, Haagmans BL (2020) Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 368:1012–1015
Rodriguez-Morales AJ, Bonilla-Aldana DK, Balbin-Ramon GJ, Rabaan AA, Sah R, Paniz-Mondolfi A, Pagliano P, Esposito S (2020) History is repeating itself: probable zoonotic spillover as the cause of the 2019 novel Coronavirus Epidemic. Infez Med 28:3–5
Rowe T, Gao G, Hogan RJ, Crystal RG, Voss TG, Grant RL, Bell P, Kobinger GP, Wivel NA, Wilson JM (2004) Macaque model for severe acute respiratory syndrome. J Virol 78:11401–11404
Saad M, Omrani AS, Baig K, Bahloul A, Elzein F, Matin MA, Selim MA, Al Mutairi M, Al Nakhli D, Al Aidaroos AY, Al Sherbeeni N, Al-Khashan HI, Memish ZA, Albarrak AM (2014) Clinical aspects and outcomes of 70 patients with Middle East respiratory syndrome coronavirus infection: a single-center experience in Saudi Arabia. Int J Infect Dis 29:301–306
Sarma P, Kaur H, Kumar H, Mahendru D, Avti P, Bhattacharyya A, Prajapat M, Shekhar N, Kumar S, Singh R, Singh A, Dhibar DP, Prakash A, Medhi B (2020) Virological and clinical cure in COVID-19 patients treated with hydroxychloroquine: a systematic review and meta-analysis. J Med Virol 92:776–785
Sia SF, Yan LM, Chin AWH, Fung K, Choy KT, Wong AYL, Kaewpreedee P, Perera RAPM, Poon LLM, Nicholls JM, Peiris M, Yen HL (2020) Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. https://doi.org/10.1038/s41586-020-2342-5
Subbarao K, McAuliffe J, Vogel L, Fahle G, Fischer S, Tatti K, Packard M, Shieh WJ, Zaki S, Murphy B (2004) Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J Virol 78:3572–3577
Subbarao K, Roberts A (2006) Is there an ideal animal model for SARS? Trends Microbiol 14:299–303
Sun J, Zhuang Z, Zheng J, Li K, Wong RL, Liu D, Huang J, He J, Zhu A, Zhao J, Li X, Xi Y, Chen R, Alshukairi AN, Chen Z, Zhang Z, Chen C, Huang X, Li F, Lai X, Chen D, Wen L, Zhuo J, Zhang Y, Wang Y, Huang S, Dai J, Shi Y, Zheng K, Leidinger MR, Chen J, Li Y, Zhong N, Meyerholz K, McCray PB, Perlman S, Zhao J (2020a) Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell. https://doi.org/10.1016/j.cell.2020.06.010
Sun SH, Chen Q, Gu HJ, Yang G, Wang YX, Huang XY, Liu SS, Zhang NN, Li XF, Xiong R, Guo Y, Deng YQ, Huang WJ, Liu Q, Liu QM, Shen YL, Zhou Y, Yang X, Zhao TY, Fan FA, Zhou YS, Qin CF, Wang YC (2020b) A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host & Microbe. https://doi.org/10.1016/j.chom.2020.05.020
Tao X, Garron T, Agrawal AS, Algaissi A, Peng BH, Wakamiya M, Chan TS, Lu L, Du L, Jiang S, Couch RB, Tseng CT (2015) Characterization and demonstration of the value of a lethal mouse model of middle east respiratory syndrome coronavirus infection and disease. J Virol 90:57–67
ter Meulen J, Bakker AB, van den Brink EN, Weverling GJ, Martina BE, Haagmans BL, Kuiken T, de Kruif J, Preiser W, Spaan W, Gelderblom HR, Goudsmit J, Osterhaus AD (2004) Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet 363:2139–2141
Vergara-Alert J, Raj VS, Muñoz M, Abad FX, Cordón I, Haagmans BL, Bensaid A, Segalés J (2017) Middle East respiratory syndrome coronavirus experimental transmission using a pig model. Transbound Emerg Dis 64:1342–1345
Vogel LN, Roberts A, Paddock CD, Genrich GL, Lamirande EW, Kapadia SU, Rose JK, Zaki SR, Subbarao K (2007) Utility of the aged BALB/c mouse model to demonstrate prevention and control strategies for severe acute respiratory syndrome coronavirus (SARS-CoV). Vaccine 25:2173–2179
World Health Organization (WHO) (2019) World Health Organization. Middle East respiratory syndrome coronavirus (MERS-CoV). https://www.who.int/emergencies/mers-cov/en/
World Health Organization (WHO) (2020a) Coronavirus disease 2019 (COVID-19) situation report. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
World Health Organization (WHO) (2020b) COVID-19 situation report-142. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200610-covid-19-sitrep-142.pdf?sfvrsn=180898cd_6
World Health Organization (WHO) (2003) Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. https://www.who.int/csr/sars/country/table2003_09_23/en/
Woolsey C, Borisevich V, Prasad AN, Agans KN, Deer DJ, Dobias NS, Heymann JC, Foster SL, Levine CB, Medina L, Melody K, Geisbert JB, Fenton KA, Geisbert TW, Cross RW (2020) Establishment of an African green monkey model for COVID-19. bioRxiv. https://doi.org/10.1101/2020.05.17.100289
Yang XH, Deng W, Tong Z, Liu YX, Zhang LF, Zhu H, Gao H, Huang L, Liu YL, Ma CM, Xu YF, Ding MX, Deng HK, Qin C (2007) Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp Med 57:450–459
Yao Y, Bao L, Deng W, Xu L, Li F, Lv Q, Yu P, Chen T, Xu Y, Zhu H, Yuan J, Gu S, Wei Q, Chen H, Yuen KY, Qin C (2014) An animal model of MERS produced by infection of rhesus macaques with MERS coronavirus. J Infect Dis 209:236–242
Yu P, Qi F, Xu Y, Li F, Liu P, Liu J, Bao L, Deng W, Gao H, Xiang Z, Xiao C, Lv Q, Gong S, Liu J, Song Z, Qu Y, Xue J, Wei Q, Liu M, Wang G, Wang S, Yu H, Liu X, Huang B, Wang W, Zhao L, Wang H, Ye F, Zhou W, Zhen W, Han J, Wu G, Jin Q, Wang J, Tan W, Qin C (2020) Age-related rhesus macaque models of COVID-19. Animal Model Exp Med 3:93–97
Zhao G, Jiang Y, Qiu H, Gao T, Zeng Y, Guo Y, Yu H, Li J, Kou Z, Du L, Tan W, Jiang S, Sun S, Zhou Y (2015) Multi-organ damage in human dipeptidyl peptidase 4 transgenic mice infected with middle east respiratory syndrome-coronavirus. PLoS ONE 10:e0145561
Zhao J, Li K, Wohlford-Lenane C, Agnihothram SS, Fett C, Zhao J, Gale MJ Jr, Baric RS, Enjuanes L, Gallagher T, McCray PB Jr, Perlman S (2014) Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci U S A 111:4970–4975
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating, and Research Team (2020) A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733
Acknowledgements
The authors would thanks Dr Tulsi Das Library, PGIMER (Institute Library).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Rights and permissions
About this article
Cite this article
Singh, A., Singh, R.S., Sarma, P. et al. A Comprehensive Review of Animal Models for Coronaviruses: SARS-CoV-2, SARS-CoV, and MERS-CoV. Virol. Sin. 35, 290–304 (2020). https://doi.org/10.1007/s12250-020-00252-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12250-020-00252-z