Abstract
The virulence properties of many pathogenic bacteria are due to proteins encoded by large gene clusters called pathogenicity islands1,2, which are found in a variety of human pathogens including Escherichia coli, Salmonella, Shigella, Yersinia, Helicobacter pylori, Vibrio cholerae, and animal and plant pathogens such as Dichelobacter nodosus and Pseudomonas syringae1,2,3. Although the presence of pathogenicity islands is a prerequisite for many bacterial diseases, little is known about their origins or mechanism of transfer into the bacterium. The bacterial agent of epidemic cholera, Vibrio cholerae , contains a bacteriophage known as cholera-toxin phage (CTXφ)4, which encodes the cholera toxin, and a large pathogenicity island called the VPI (for V. cholerae pathogenicity island)5 which itself encodes a toxin-coregulated pilus that functions as a colonization factor6 and as a CTXφ receptor4. We have now identified the VPI pathogenicity island as the genome of another filamentous bacteriophage, VPIφ. We show that VPIφ is transferred between V. cholerae strains and provide evidence that the TcpA subunit of the toxin-coregulated type IV pilus is in fact a coat protein of VPIφ. Our results are the first description of a phage that encodes a receptor for another phage and of a virus–virus interaction that is necessary for bacterial pathogenicity.
Similar content being viewed by others
Main
Cholera is an ancient and life-threatening epidemic disease that occurs worldwide7,8. Virulent and epidemic strains of V. cholerae require two genetic elements to cause disease, CTXφ4 and VPI5. CTXφ encodes the cholera toxin responsible for the severe secretory diarrhoea characteristic of the disease4. The VPI (Fig. 1) is required for the emergence of V. cholerae as it contains the toxin-coregulated pilus (TCP) gene cluster which encodes a type-IV pilus that functions both as an essential colonization factor6,9 and as a CTXφ receptor4. VPI has many features of bacterial pathogenicity islands (PAIs): it is large (∼40 kilobases), contains genes associated with virulence, regulation and mobility, is inserted into a single chromosomal site (att site) adjacent to a tRNA-like gene, and it has a different G+ C content compared with the host chromosome4,5,6,10,11,12,13.
Arrows above genes indicate the PCR primers used and are identified by the number of the KAR primer series. Black bars, regions targeted for PCR; grey bars, common chromosomal flanking DNA; checked regions at each end of VPI denote att sites; dotted region (left) in the VPI represents the defective transposase (xtn). The aphA -3 gene encoding Km resistance is inserted into a Bam HI site between xtn and aldA.
We previously identified one V. cholerae strain (E9120) that has lost the VPI (ref. 5). This strain contains ctx genes, suggesting that it previously had the VPI to allow infection by CTXφ, and has an altered att site, indicating that the VPI had been excised. As the VPI contains a transposase-like gene and a phage-like integrase gene5,13, we investigated other VPI genes using BLAST14 to find ones whose predicted protein products share homology with phage or viral proteins. TagE has 30% identity and 55% similarity over 188 amino acids to Orf16 of Staphylococcus aureus bacteriophage φ (Genbank accession number, AB009866); OrfZ has 26% identity and 41% similarity over 97 amino acids to an ‘early’ protein of rat cytomegalovirus (Genbank accession number, U62396); and OrfV has 32% identity and 47% similarity over 79 amino acids to the ‘enhancin’ protein of Lymantria dispar nucleopolyhedrovirus (Genbank accession number, AF019970). Even TcpA from the El Tor strain N16961 shows homology (31% identity, 48% similarity over 82 amino acids) to the product of a 769-amino-acid Orf from TT virus (Genbank accession number, AB011490). Taken together, these results indicated that the VPI could be the genome of a phage.
We tested this idea by examining phage preparations of V. cholerae strains N16961 and 395 for genes at the ends and centre of the VPI. By using the polymerase chain reaction (PCR) and sequencing, we found orf1, tcpA, toxT and int genes in both phage preparations (Fig. 1, 2 and Table 1), but no PCR products were amplified when primers targeted regions immediately outside the VPI, nor for ompT (which encodes an outer membrane protein) (Fig. 2) or rfaD (involved in lipopolysaccharide synthesis), which are also outside the VPI. A phage preparation derived from a VPI-negative strain prepared under identical conditions was also negative for the chromosomal ompT and rfaD, as well as for orf1, tcpA, toxT and int genes. As expected, PCR analysis of the preparations identified ctxB and zot genes on CTXφ. Finding VPI genes in phage preparations after treatment with DNase and RNase indicated that the DNA was protected, presumably by a protein coat, and that this element was probably a bacteriophage (designated VPIφ).
Phage DNA from strains N16961 and 395 was sensitive to digestion with S1 nuclease (specific for single-stranded DNA) and S1-treated phage DNA gave no PCR products. The same phage DNA was resistant to digestion with restriction endonucleases specific for double-stranded DNA and yielded VPI PCR products after digestion. Southern-blot analysis on phage DNA from DK238 (see below) and CVD110 using tcpA forward and reverse primers revealed that the phage DNA hybridized only with the primer that could hybridize to the positive strand (Fig. 3). Thus, VPIφ, like CTXφ (ref. 4), contains positive, single-strand DNA as its genome.
To determine whether VPIφ has a plasmid replicative form (RF) in the cell, we analysed plasmid DNA from N16961 and 395 for VPI genes. RF preparations were sensitive to digestion with double-strand-specific Pvu II (which has a site in tcpA) as Pvu II-digested RF preparations failed to generate PCR fragments using tcpA primers (Fig. 4a ), but were resistant to digestion with S1 nuclease and could still generate VPI PCR products after treatment with S1. PCR analysis revealed no chromosomal contamination as ompT and rfaD sequences were not amplified (Table 1). Southern-blot hybridization of RF preparations confirmed the presence of a VPIφ replicative form. Hae III-digested RF from DK238 and CVD110, as well as N16961 chromosomal DNA, hybridized with a tcpA PCR product probe, whereas only chromosomal DNA hybridized with an ompT probe (Fig. 4b, c). These results indicate that, like other filamentous phages, VPIφ forms a double-stranded-DNA plasmid replicative form in the cell.
a, PCR analysis of the RF using tcpA primers KAR24 and KAR25. Note that a tcpA product is not generated by using Pvu II-digested RF as template, whereas RF treated with S1 nuclease (RF/S1) generates a PCR product. N16961 chromosome was used as a control template. b, Southern-blot hybridization of Hae III-digested RF from DK238 and CVD110 probed with a tcpA PCR product; c, Southern-blot hybridization of Hae III-digested RF probed with an ompT PCR product shows hybridization only to the N16961 chromosomal control.
The VPIφ genome of a spontaneously streptomycin-resistant (Str) strain (N16961) was marked with the aphA -3 gene, which encodes resistance to kanamycin (Km) and neomycin (Neo), creating strain DK238. aphA -3 was inserted between aldA and xtn (Fig. 1), a region encoding a putative non-functional transposase5. We used whole cells and cell-free phage preparations from donor DK238 to determine whether VPIφ could be transferred into the non-toxigenic VPI-negative strain DK236 (serogroup O10, nalidixic acid (Nal) resistant). After 1 hour of incubation at 37 °C of 3× 107 DK236 recipient cells with either 3× 107 donor DK238 cells or phage preparations from 2× 107 DK238 cells, cultures were plated onto agar containing Nal/Neo. The use of a Nal-sensitive donor and Nal-resistant recipient enabled us to identify Nal-resistant tranductants that had acquired aphA -3 from VPIφ. With DK238 cells, we obtained 1.3 × 104 transductants, suggesting that, on average, about 0.04% of recipient cells were transduced under these conditions; with phage preparations from 2× 107 donor cells, we obtained >3 × 108 transductants per 3× 107 recipients, indicating that VPIφ made by one donor cell gives rise to at least 15 transductants. The successful transfer of the neomycin-resistance marker with cell-free DNase-treated phage preparations indicated that transduction, rather than conjugation, was the mechanism of DNA transfer.
Transfer of VPIφ from donor to recipient was confirmed for one transductant, DK239, by using O antigen agglutination, antibiotic-susceptibility testing, ribotyping, colony hybridization with aphA -3 and tcpA probes, and PCR analysis of aphA -3, tcpA, toxT and int genes (Table 1). PCR on DK239 using primers for the right VPI junction showed that the VPI had integrated into the same site as wild-type VPI-positive strains. PCR on phage preparations from DK239 indicates that, although DK239 produces VPIφ, CTXφ was not acquired (Table 1).
Not all V. cholerae strains could act as donors or recipients of VPIφ. Although we detected VPIφ particles in phage preparations of classical strain 395, no VPIφ was transferred when a derivative of 395 (also marked by aphA -3 in the VPI) was used as donor, suggesting that this strain is unable to transfer VPIφ efficiently, or that different conditions are needed. With N16961 as donor, transfer occurred into the O10 strain DK236 but not into the non-toxigenic VPI-negative strain DK237 (serogroup O1). As both DK236 and DK237 have a vacant att site5, these strains may differ in their ability to acquire VPIφ, possibly explaining the limited number of toxigenic serogroups. Epidemic and pandemic cholera is associated with O1 and O139 strains. Although we have shown that VPI+ non-O1/non-O139, potentially toxigenic strains can be created in vitro, VPI+CT+ O1 strains may have an advantage in vivo or in the environment over VPI+CT+ non-O1 strains, explaining the predominance of O1 strains in epidemic and pandemic disease. Our results highlight the potential for serogroups of V. cholerae other than O1 and O139 to acquire the VPI and become pathogenic and even epidemic and pandemic strains.
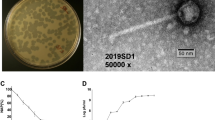
Concentrated phage preparations of several VPI-positive V. cholerae strains were viewed under electron microscopy. Immunoelectron microscopy of preparations from strain 395 using rabbit anti-TcpA antibodies revealed many gold particles bound to parallel bundles of VPIφ particles (Fig. 5a). El Tor strain CVD110, which has the ctxA, zot, ace and orfU genes deleted15, cannot produce CTXφ or CTXφ-encoded genes, but does contain VPIφ genes (Table 1). Our CVD110 preparation contained many phage particles, some of which formed a ‘braided’ network of filaments, presumably representing VPIφ (Fig. 5b). Immunoelectron microscopy of phage preparations from El Tor strain N16961, like 395, revealed numerous gold particles bound to filamentous phage (Fig. 5c ).
a, Phage preparations from 395 incubated with rabbit anti-TcpA antibody and 10-nm colloidal gold-conjugated goat anti-rabbit IgG (original magnification, ×50,000; scale bar, 100 nm); b, phage preparations from CVD110 (magnification, ×100,000; scale bar, 50 nm). c, Phage preparations from N16961 incubated with rabbit anti-TcpA antibody and 10-nm colloidal gold-conjugated goat anti-rabbit IgG (magnification ×120,000; scale bar, 50 nm).
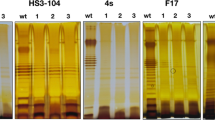
As VPI is a bacteriophage and the structure of type-IV pili resembles that of a filamentous bacteriophage16, we investigated whether the TCP pilin subunit, TcpA, could be a VPIφ coat protein. TCP2 is a derivative of strain 395 with a large internal deletion in the tcpA gene that prevents it from producing TCP17. No VPIφ genes were detected in phage preparations from TCP2, indicating that TcpA is required for VPIφ production; in contrast, CTXφ genes were evident (Table 1and Fig. 2). In immunoprecipitation experiments on these phage preparations, rabbit anti-TcpA-peptide antibodies and mouse anti-rabbit agarose beads bound VPIφ, allowing selective removal of the complex. PCR analysis on N16961 and 395 immunoprecipitates revealed VPIφ-encoded genes (Fig. 6). We demonstrated antibody specificity for TcpA and VPIφ by the lack of PCR products in similar experiments done without anti-TcpA antibodies (Table 2). PCR on phage preparations from an independent tcpA mutant, RT4032, which contains an in-frame (non-polar) deletion in tcpA, also failed to generate VPI genes; however, the tcpA mutants TCP2 and RT4032 could be complemented by supplying tcpA on a plasmid (pRT198) because PCR detected VPI genes in phage preparations of both transformants (Fig. 7). These results together support the idea that TcpA is an important coat protein of VPIφ.
a, PCR done with tcpA primers KAR24 and KAR82; b, PCR done with orf1 primers; c, PCR done with rfaD primers. Note the absence of a tcpA and orf1 PCR product in phage preparations from RT4032 (owing to its inability to make VPIφ) and the presence of products when RT4032 is complemented with pRT198 supplying tcpA.
Our findings help explain how non-pathogenic bacteria can become pathogens. Many bacterial pathogens contain clusters of genes that encode virulence factors responsible for inducing disease. We have shown that the large cluster of genes essential for the epidemic properties of V. cholerae originates from viral (bacteriophage) DNA that has become incorporated into the bacterial chromosome, and that transfer of the unusually large filamentous bacteriophage VPIφ confers new virulence on the recipient. Type IV pili like TCP are expressed by a variety of human and animal pathogens, including enteropathogenic E. coli, Neisseria gonorrhoeae, P. aeruginosa and D. nodosus18,19,20,21, and may also have a bacteriophage origin and transferable genes that endow the bacterium with virulence factors. It is not understood how TCP serves as both a bacteriophage and colonization factor, particularly as it has not been shown to bind directly to intestinal tissue or to cultured epithelial cells. The type-IV pilus of enteropathogenic E. coli, BFP, which shares significant homology with TCP, was recently reported to function in colonization by mediating bacteria-to-bacteria adherence, thereby increasing the bacterial mass that colonizes the intestine22. TCP likewise mediates interbacterial adherence (autoagglutination is a standard assay for this structure), so released VPIφ/TCP pili might help colonization by serving as a bridge between phage/pili bound to different bacteria. TCP is also the receptor for CTXφ (ref. 4), creating a situation in which one phage serves as the receptor for a second phage in a sequential infection process that results in bacterial virulence. Our results highlight the potential contribution of virulence-conferring phage (‘pathophage’) in the emergence of pathogens, now and in the future.
Methods
Bacterial strains and plasmids. Strain 395 is a representative of the sixth pandemic clone (classical biotype) of V. cholerae; strain N16961 is an isolate from the current seventh (‘El Tor’) pandemic which began in 1961 in Indonesia. CVD110 is a derivative of a seventh pandemic strain (E7946) which is deleted in its ctxA, zot, ace and orfU genes15. The environmental isolates DK236 and DK237 are non-toxigenic VPI-negative serogroup O10 and O1 strains, respectively, and are Nal-resistant owing to selection of a spontaneous resistant mutant. To construct DK238, the xtn–aldA region of the chromosomally integrated N16961 VPI was amplified by PCR with primers KAR166 (located in orf1) and KAR167 (located in aldA). This 1.8-kb fragment was ligated into pGEM-T, creating pDK40. This plasmid was digested with Bam Hl and the aphA -3 gene (encoding Km/Neo resistance) was inserted, creating pDK42, which was digested with Sph I/Sac I; the xtn–aldA ::aphA -3 fragment was ligated into the suicide vector pCVD442, creating pDK43. Plasmid pDK43 was used in an allelic exchange procedure to introduce the aphA -3-containing fragment into the homologous region of the N16961 chromosome, creating strain DK238. Strain TCP2 is derived from 395, has amino-acid residues 119–154 deleted from its tcpA gene and does not produce the TCP structure17. RT4032 (from R. Taylor) is an in-frame tcpA deletion mutant of 395 in which the codons encoding amino acids +1 of mature TcpA through the TAA stop codon and one additional T nucleotide are removed. Plasmid pRT198 is a pBR322-based plasmid with a 2-kb Hin dIII fragment containing tcpA of 395 cloned into the Hin dIII site of pBR322.
PCR and sequencing. PCR was performed as described23 under the following conditions: denature at 96 °C for 3 min; annealing, 48 °C, 30 s; extension, 72 °C, 2 min for 1 cycle, then denature 96 °C, 30 s; annealing, 48 °C, 30 s; extension, 72 °C, 2 min for 30 cycles. Primers used to determine the presence and sequence of V. cholerae genes were: orf1, KAR96, 5′-TGCTACTTACCCAATGGCAC-3′ and KAR97, 5′-GAGCCAGGCTTATTTGGGCG-3′; tcpA, KAR24, 5′-AAAACCGGTCAAGAGGG-3′ and KAR25, 5′-CAAAAGCTACTGTGAATGG-3′ for seventh pandemic strains and KAR82, 5′-CAAATGCAACGCCGAATGG-3′ for sixth pandemic strains; toxT, KAR90, 5′-ATAACTTTACGTGGATGGC-3′ and KAR91, 5′-AAAATCAGTGATACAATCG-3′; int, KAR22, 5′-GATAAAGAGATCAAAGCC-3′ and KAR23, 5′-ATCTGCTTCCATGTGGG-3′. Additional primers included KAR94, 5′-TATGATACTGAAAACACCTC-3′ and KAR95, 5′-GATGCTAACAGCAGAGCATA-3′ (outside left VPI junction); KAR85, 5′-CGCCTGCGAACCGACACGC-3′, and KAR86, 5′-GCAGCAAGCCTCCACTCCG-3′ (outside the right VPI junction); K898, 5′-GAATTCTGTCGGGTTGTAATCCTG-3′ and K643, 5′-GCCATACTCAGCATATACAC-3′ (ompT); K371, 5′-CGGGATCCGAGCTCATTACCTACACTAGTG-3′ and K369, 5′-CGGGATCCGACAGGCTATAATGCGTGCAAC-3′ (rfaD); KAR161, 5′-AAAATTCCTTGACGAATACC-3′ and KAR162, 5′-TTGCTTCTCATCATCGAACC-3′ (ctxB); KAR166, 5′-GACAGGATTACTGAGATATCTG-3′ and KAR167, 5′-AACCAAGGTGAGGTTTGTACC-3′ (xtn-aldA region). Primers were synthesized using an Applied Biosystems DNA synthesizer and sequenced with a Taq Dye-Terminator kit (Perkin-Elmer) and an automated 373A DNA sequencer (Applied Biosystems).
Isolation of phage and replicative form. Isolation of phage from V. cholerae was done essentially as described24. In brief, 1-litre Luria broth cultures were grown overnight at either 30 °C (395) or 37 °C (N16961). Cultures were centrifuged twice at 10,000 g and the supernatant was passed through a 0.45-µm low-protein-binding filter. DNase I and RNase I (Boehringer Mannheim) were added to the filtrate at a final concentration of 1 µg ml−1 and incubated at room temperature for 3 h. NaCl and PEG 8000 were added to a final concentration of 1 M and 10% w/v, respectively, and the mixture was left to precipitate overnight at 4 °C. The supernatant was centrifuged at 11,000 g for 20 min and the pellet resuspended in 4 ml of SM buffer. PEG was removed with an equal volume of chloroform. This supernatant was layered onto a CsCl2 step gradient consisting of 2 ml each of CsCl2 in SM buffer ( d = 1.7, 1.5 and 1.45). After centrifugation at 25,000 r.p.m. for 2 h in an SW41 rotor (Beckman), the lower phage band (1.45 ⩽ d ⩽1.5) was extracted and dialysed against two changes of TM buffer. The phages were further concentrated by the addition of PEG 8000 (10% w/v) and placed on ice for 2 h. The phage preparation was centrifuged at 14,000 g for 20 min and the resulting pellet (containing phage particles) was resuspended in 100 µl of SM buffer. PEG was removed with an equal volume of chloroform. 5 µl of the phage preparation was used for PCR.
The replicative form was isolated from 1 litre of Luria broth culture as described24.
Although filamentous phage do not lyse the cell or have a ‘burst size’, we calculated the approximate number of VPIφ and CTXφ released per cell and the RF copy number. The number of phage released was calculated by determining the amount of phage DNA (from its absorbance) in 1 litre of overnight culture of DK238 and CVD110 containing ∼1012 cells. As strain DK238 is positive for VPIφ and CTXφ and CVD110 is positive for only VPIφ, the difference in amount (in µg) between the two strains should reflect the number of CTXφ genomes (7 kb) and the balance the number of VPIφ genomes (40 kb) released per cell. We estimate that 280 and 200 µg l−1 of ssDNA is present in a phage preparation from 1-litre cultures of DK238 and CVD110, respectively. If 1 µg of 1-kb ssDNA contains 1.8 × 1012 molecules, then 1 µg of 7-kb ssDNA contains 2.6 × 1011 CTXφ molecules and 80 µg contains 2× 1013 molecules, so 2× 1013 molecules/1012 cells indicates that an average of 20 CTXφ are made per cell during overnight culture. Likewise, for VPIφ we calculate that an average of 9 VPIφ are produced per cell. Our calculations all assume that no other phage (or RF) is produced and that DNA extraction is 100% efficient, which is unlikely, so values could be underestimates.
We estimated that 57 and 39 µg l−1 of dsDNA are present in RF preparations from 1-litre cultures of DK238 and CVD110, respectively, which yields an average CTXφ RF copy number of 2 per cell after overnight culture; the average VPIφ RF copy number is similarly estimated as 1 per cell.
Immunoprecipitation. DNase- and RNase-treated phage preparations were incubated with rabbit anti-TcpA peptide antibody (1:10,000) at 37 °C and with vigorous shaking for 1 h. Mouse anti-rabbit IgG (whole molecule) agarose beads (Sigma) were added and the reaction was incubated at 4 °C overnight with gentle rotation. After several low-speed centrifugations and washings in deionized water, the pellet was resuspended in deionized water.
Electron microscopy. Phage preparations of CVD110 and 395 were placed on a carbon–formvar-coated 300-mesh copper grid (Electron Sciences) for 2 min then negatively stained with 1.5% phosphotungstic acid for 1 min and analysed by electron microscopy. In addition, equal volumes of 395 and N16961 phage preparations were separately incubated with rabbit anti-peptide TcpA antibody (1:10,000) at 37 °C for 20 min, after which this suspension was placed on a grid for 5 min, and 10 µl of 10-nm colloidal gold-conjugated goat anti-rabbit IgG (ICN Biomedicals) was added before negative staining.
References
Lee, C. A. Pathogenicity islands and evolution of bacterial pathogens. Infect. Agent. Dis. 5, 1–7 ( 1996).
Hacker, J., Blum-Oehler, G., Muhldorfer, I. & Tschape, H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23, 1089–1097 (1997).
Huang, H. C., Lin, R. H., Chang, C. J., Collmer, A. & Deng, W. L. The complete hrp gene cluster of Pseudomonas syringae pv. syringae 61 includes two blocks of genes required for harpinPss secretion that are arranged colinearly with Yersinia ysc homologs. Mol. Plant. Microbe Interns 8, 733–746 (1995).
Waldor, M. K. & Mekalanos, J. J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272, 1910–1914 (1996).
Karaolis, D. K. R.et al. AVibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl Acad. Sci. USA 95, 3134–3139 (1998).
Taylor, R. K., Miller, V. L., Furlong, D. B. & Mekalanos, J. J. The use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl Acad. Sci. USA 84, 2833–2837 ( 1987).
Politzer, R. Cholera. Monogr. Ser. 43 (Geneva, World Health Organization, (1959).
Kaper, J. B., Morris, J. G. J & Levine, M. M. Cholera. Clin. Microbiol. Rev. 8, 48–86 (1995).
Herrington, D. A. et al. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168, 1487–1492 (1988).
DiRita, V. J., Parsot, C., Jander, G. & Mekalanos, J. J. Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl Acad. Sci. USA 88, 5403–5407 (1991).
Carroll, P. A., Tashima, K. T., Rogers, M. B., DiRita, V. J. & Calderwood, S. B. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol. Microbiol. 25, 1099–1111 (1997).
Häse, C. C. & Mekalanos, J. J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae . Proc. Natl Acad. Sci. USA 95, 730– 734 (1998).
Kovach, M. E., Shaffer, M. D. & Peterson, K. M. Aputative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae . Microbiology 142, 2165– 2174 (1996).
Altschul, S. F.et al . Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Michalski, J., Galen, J. E., Fasano, A. & Kaper, J. B. CVD110, an attenuated Vibrio cholerae O1 El Tor live oral vaccine strain. Infect. Immun. 61, 4462–4468 ( 1993).
Hobbs, M. & Mattick, J. S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10, 233– 243 (1993).
Taylor, R. K., Shaw, C. E., Peterson, K. M., Spears, P. & Mekalanos, J. J. Safe, live Vibrio cholerae vaccines? Vaccine 6, 151– 154 (1988).
Dalrymple, B. & Mattick, J. S. An analysis of the organization and evolution of type 4 fibrial (MePhe) subunit proteins. J. Mol. Evol. 25, 261–269 ( 1987).
Shaw, C. E. & Taylor, R. K. Vibrio cholerae O395 tcpA pilin gene sequence and comparison of predicted protein structural features to those of type 4 pilins. Infect. Immun. 58, 3042–3049 (1990).
Patel, P.et al. Shared antigenicity of type 4 pilins expressed by Pseudomonas aeruginosa, Moraxella bovis, Neisseria gonorrhoeae, Dichelobacter nodosus, and Vibrio cholerae. Infect. Immun. 59, 4674–4676 ( 1991).
Girón, J. A., Ho, A. S. Y. & Schoolnick, G. K. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254, 710– 713 (1991).
Hicks, S., Frankel, G., Kaper, J. B., Dougan, G. & Phillips, A. D. Role of intimin and bundle-forming pili in enteropathogenic Escherichia coli adhesion to pediatric intestinal tissue in vitro. Infect. Immun. 66, 1570–1578 (1998).
Saiki, R. K.et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239, 487– 491 (1988).
Maniatis, T., Fritsch, E. F. & Sambrook, J. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, (1982 ).
Acknowledgements
We thank our colleagues at the University of Maryland for their support, particularly D. Hone for discussions, T. Agin, J. Michalski and M. Boyanapalli for technical assistance, and R. T. Milanich for help with prepAring the figures; and R. Taylor for RT4032, pRT198 and anti-peptide 6 TcpA antibody. This work was supported by grants from the NIH and Department of Veterans Affairs. D.K.R.K. is a recipient of a Burroughs Wellcome Fund Career Award in the Biomedical Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Karaolis, D., Somara, S., Maneval, D. et al. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399, 375–379 (1999). https://doi.org/10.1038/20715
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/20715
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.