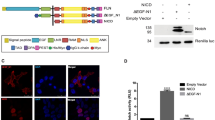
Abstract
Notch signalling controls growth, differentiation and patterning during normal animal development1,2; in humans, aberrant Notch signalling has been implicated in cancer and stroke3,4. The mechanism of Notch signalling is thought to require cleavage of the receptor in response to ligand binding5, movement of the receptor's intracellular domain to the nucleus6,7, and binding of that intracellular domain to a CSL (for CBF1, Suppressor of Hairless, LAG-1)8,9 protein. Here we identify LAG-3, a glutamine-rich protein that forms a ternary complex together with the LAG-1 DNA-binding protein10 and the receptor's intracellular domain. Receptors with mutant ankyrin repeats that abrogate signal transduction are incapable of complex formation both in yeast and in vitro. Using RNA interference, we find that LAG-3 activity is crucial in Caenorhabditis elegans for both GLP-1 and LIN-12 signalling. LAG-3 is a potent transcriptional activator in yeast, and a Myc-tagged LAG-3 is predominantly nuclear in C. elegans. We propose that GLP-1 and LIN-12 promote signalling by recruiting LAG-3 to target promoters, where it functions as a transcriptional activator.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kimble, J. & Simpson, P. The LIN-12/Notch signalling pathway and its regulation. Annu. Rev. Cell Dev. Biol. 13, 333–361 (1997).
Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. Notch signalling: cell fate control and signal integration in development. Science 284, 770– 776 (1999).
Ellisen, L. W. et al. TAN-1, the human homolog of the Drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66, 649–661 ( 1991).
Joutel, A. et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature 383, 707–710 (1996).
Schroeter, E. H., Kisslinger, J. A. & Kopan, R. Notch1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 (1998).
Struhl, G. & Adachi, A. Nuclear access and action of Notch in vivo. Cell 93, 649– 660 (1998).
Lecourtois, M. & Schweisguth, F. Indirect evidence for Delta-dependent intracellular processing of notch in Drosophila embryos. Curr. Biol. 8, 771–774 ( 1998).
Roehl, H., Bosenberg, M., Blelloch, R. & Kimble, J. Roles of the RAM and ANK domains in signalling by the C. elegans GLP-1 receptor. EMBO J. 15, 7002– 7012 (1996).
Tamura, K. et al. Physical interaction between a novel domain of receptor Notch and the transcription factor RBP-Jκ/Su(H). Curr. Biol. 5, 1416–1423 (1995).
Christensen, S., Kodoyianni, V., Bosenberg, M., Friedman, L. & Kimble, J. lag-1, a gene required for lin-12 and glp-1 signalling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su(H). Development 122, 1373–1383 ( 1996).
Roehl, H. & Kimble, J. Control of cell fate in C. elegans by a GLP-1 peptide consisting primarily of ankyrin repeats. Nature 364, 632–635 ( 1993).
Greenwald, I. & Seydoux, G. Analysis of gain-of-function mutations of the lin-12 gene of Caenorhabditis elegans. Nature 346, 197–199 ( 1990).
Kodoyianni, V., Maine, E. M. & Kimble, J. Molecular basis of loss-of-function mutations in the glp-1 gene of Caenorhabditis elegans. Mol. Biol. Cell 3, 1199–1213 ( 1992).
Mango, S. E., Maine, E. M. & Kimble, J. Carboxy-terminal truncation activates glp-1 protein to specify vulval fates in Caenorhabditis elegans. Nature 352, 811–815 ( 1991).
Austin, J. & Kimble, J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans . Cell 51, 589–599 (1987).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Greenwald, I. S., Sternberg, P. W. & Horvitz, H. R. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 34, 435 –444 (1983).
Berry, L. W., Westlund, B. & Schedl, T. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development 124, 925– 936 (1997).
Mitchell, P. J. & Tjian, R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245, 371–378 ( 1989).
Hicks, G. R. & Raikhel, N. Y. Protein import into the nucleus: an integrated view. Annu. Rev. Cell Dev. Biol. 11, 155–188 (1995).
Diederich, R. J., Matsuno, K., Hing, H. & Artavanis-Tsakonas, S. Cytosolic interaction between deltex and Notch ankyrin repeats implicates deltex in the Notch signalling pathway. Development 120, 473–481 (1994).
Kopan, R., Nye, J. S. & Weintraub, H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development 120, 2385– 2396 (1994).
Jarriault, S. et al. Signalling downstream of activated mammalian Notch. Nature 377, 355–358 ( 1995).
Kato, H. et al. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development 124, 4133 –4141 (1997).
Smoller, D. et al. The Drosophila neurogenic locus mastermind encodes a nuclear protein unusually rich in amino acid homopolymers. Genes Dev. 4, 1688–1700 ( 1990).
Bartel, P. L. & Fields, S. (eds) The Yeast Two-Hybrid System (Oxford Univ. Press, New York, 1997).
Kraemer, B. et al. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol. 9, 1009–1018 ( 1999).
The C. elegans Sequencing Consortium., Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018 (1998); erratum ibid 283, 35 (1999); erratum ibid 283, 2103 (1998).
Subramaniam, K. & Seydoux, G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126, 4861–4871 (1999).
Crittenden, S. L. & Kimble, J. in Cell: A Laboratory Manual (eds Spector, D., Goldman, R. & Leinwand, L.) 108.101 –108.109 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 1998).
Acknowledgements
We acknowledge A. Puoti for providing the plasmid form of the cDNA library; R. Sternglanz, S. M. Hollenberg and A. Grimson for yeast strains and plasmids; and A. Steinberg and L. Vanderploeg for help with the illustrations. A.G.P. is an Howard Hughes Medical Institute predoctoral fellow. J.K. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Petcherski, A., Kimble, J. LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature 405, 364–368 (2000). https://doi.org/10.1038/35012645
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35012645
This article is cited by
-
NACK and INTEGRATOR act coordinately to activate Notch-mediated transcription in tumorigenesis
Cell Communication and Signaling (2021)
-
Transcription factor Ptf1a in development, diseases and reprogramming
Cellular and Molecular Life Sciences (2019)
-
Critical roles of NOTCH1 in acute T-cell lymphoblastic leukemia
International Journal of Hematology (2011)
-
Structural and mechanistic insights into cooperative assembly of dimeric Notch transcription complexes
Nature Structural & Molecular Biology (2010)
-
Cooperativity in macromolecular assembly
Nature Chemical Biology (2008)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.