Abstract
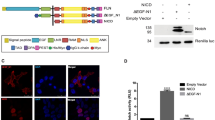
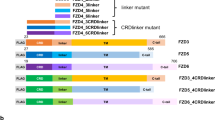
Proteins encoded by the fringe family of genes are required to modulate Notch signalling in a wide range of developmental contexts. Using a cell co-culture assay, we find that mammalian Lunatic fringe (Lfng) inhibits Jagged1-mediated signalling and potentiates Delta1-mediated signalling through Notch1. Lfng localizes to the Golgi, and Lfng-dependent modulation of Notch signalling requires both expression of Lfng in the Notch-responsive cell and the Notch extracellular domain. Lfng does not prevent binding of soluble Jagged1 or Delta1 to Notch1-expressing cells. Lfng potentiates both Jagged1- and Delta1-mediated signalling via Notch2, in contrast to its actions with Notch1. Our data suggest that Fringe-dependent differential modulation of the interaction of Delta/Serrate/Lag2 (DSL) ligands with their Notch receptors is likely to have a significant role in the combinatorial repertoire of Notch signalling in mammals.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. Notch signalling: cell fate control and signal integration in development. Science 284, 770-776 (1999).
Kuroda, K. et al. Delta-induced Notch signalling mediated by RBP-J inhibits MyoD expression and myogenesis. J. Biol. Chem. 274, 7238–7244 (1999).
Jarriault, S. et al. Delta-1 activation of notch-1 signalling results in HES-1 transactivation. Mol. Cell. Biol. 18, 7423 –7431 (1998).
Li, L. et al. The human homolog of rat Jagged1 expressed by marrow stroma inhibits differentiation of 32D cells through interaction with Notch1. Immunity 8, 43–55 ( 1998).
Lindsell, C. E., Shawber, C. J., Boulter, J. & Weinmaster, G. Jagged: a mammalian ligand that activates Notch1. Cell 80, 909–917 (1995).
Nofziger, D., Miyamoto, A., Lyons, K. M. & Weinmaster, G. Notch signalling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development 126, 1689– 1702 (1999).
Luo, B., Aster, J. C., Hasserjian, R. P., Kuo, F. & Sklar, J. Isolation and functional analysis of a cDNA for human Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol. Cell. Biol. 17, 6057–6067 (1997).
Shawber, C., Boulter, J., Lindsell, C. E. & Weinmaster, G. Jagged2: a serrate-like gene expressed during rat embryogenesis. Dev. Biol. 180, 370–376 (1996).
Lindsell, C. E., Boutler, J., diSibio, G., Gossler, A. & Weinmaster, G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol. Cell. Neurosci. 8, 14– 27 (1996).
Blair, S. S. Limb development: Marginal fringe benefits. Curr. Biol. 7, R686–R690 (1997).
Irvine, K. D. Fringe, Notch, and making developmental boundaries. Curr. Opin. Genet. Dev. 9, 434–441 ( 1999).
Panin, V. M., Papayannopoulos, V., Wilson, R. & Irvine, K. D. Fringe modulates Notch-ligand interactions. Nature 387, 908–912 (1997).
Fleming, R. J., Gu, Y. & Hukriede, N. A. Serrate-mediated activation of Notch is specifically blocked by the product of the gene fringe in the dorsal compartment of the Drosophila wing imaginal disc. Development 124, 2973 –2981 (1997).
Johnston, S. H. et al. A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development 124 , 2245–2254 (1997).
Cohen, B. et al. Fringe boundaries coincide with Notch-dependent patterning centres in mammals and alter Notch-dependent development in Drosophila. Nature Genet. 16, 283–288 (1997).
May, W. A. et al. EWS/FLI 1-induced manic fringe renders NIH3T3 cells tumorigenic . Nature Genet. 17, 495– 497 (1997).
Wu, J. Y., Wen, L., Zhang, W. J. & Rao, Y. The secreted product of Xenopus gene lunatic fringe, a vertebrate signalling molecule. Science 273, 355–358 ( 1996).
Laufer, E. et al. Expression of Radical fringe in limb-bud ectoderm regulates apical ectodermal ridge formation. Nature 386, 366–373 (1997).
Rodriguez-Esteban, C. et al. Radical fringe positions the apical ectodermal ridge at the dorsoventral boundary of the vertebrate limb. Nature 386, 360–366 (1997).
Evrard, Y. A., Lun, Y., Aulehla, A., Gan, L. & Johnson, R. L. lunatic fringe is an essential mediator of somite segmentation and patterning. Nature 394, 377– 381 (1998).
Barrantes, I. B. et al. Interaction between Notch signalling and Lunatic fringe during somite boundary formation in the mouse. Curr. Biol. 9, 470–480 (1999).
Zhang, N. & Gridley, T. Defects in somite formation in lunatic fringe-deficient mice. Nature 394, 374– 377 (1998).
Shen, J. et al. Skeletal and CNS defects in Presenilin-1-deficient mice. Cell 89, 629–639 ( 1997).
Wong, P. C. et al. Presenilin1 is required for Notch1 and DII1 expression in the paraxial mesoderm. Nature 387, 288– 292 (1997).
Irvine, K. D. & Wieschaus, E. Fringe, a boundary-specific signalling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell 79, 595– 606 (1994).
Yuan, Y. P., Schultz, J., Mlodzik, M. & Bork, P. Secreted fringe-like signalling molecules may be glycosyltransferases. Cell 88, 9–11 (1997).
Shima, D. T., Haldar, K., Pepperkok, R., Watson, R. & Warren, G. Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J. Cell. Biol. 137, 1211–1228 (1997).
Waters, M. G., Clary, D. O. & Rothman, J. E. A novel 115-kD peripheral membrane protein is required for intercisternal transport in the Golgi stack. J. Cell. Biol. 118, 1015–1026 ( 1992).
Shawber, C. et al. Notch signalling inhibits muscle cell differentiation through a CBF1-independent pathway. Development 122, 3765–3773 (1996).
Hukriede, N. A., Gu, Y. & Fleming, R. J. A dominant-negative form of Serrate acts as a general antagonist of Notch activation. Development 124, 3427–3437 (1997).
Panin, V. M. & Irvine, K. D. Modulators of Notch signalling . Semin. Cell. Dev. Biol. 9, 609– 617 (1998).
Shimizu, K. et al. Mouse Jagged1 physically interacts with Notch2 and other Notch receptors. Assessment by quantitative methods. J. Biol. Chem. 274, 32961–32969 (1999).
Sestan, N., Artavanis-Tsakonas, S. & Rakic, P. Contact-dependent inhibition of cortical neurite growth mediated by notch signalling. Science 286, 741–746 (1999).
Qi, H. et al. Processing of the Notch ligand Delta by the metalloprotease Kuzbanian . Science 283, 91–94 (1999).
Wang, S. et al. Notch receptor activation inhibits oligodendrocyte differentiation . Neuron 21, 63–75 (1998).
Varnum-Finney, B. et al. The Notch ligand, Jagged-1, influences the development of primitive hematopoietic precursor cells. Blood 91, 4084–4091 (1998).
Weinmaster, G. The ins and outs of notch signalling. Mol. Cell. Neurosci. 9, 91–102 (1997).
Moloney, D. J. et al. Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidernal growth factor-like modules. J. Biol. Chem. 275, 9604–9611 (2000).
Kusumi, K. et al. The mouse pudgy mutation disrupts Delta homologue Dll3 and initiation of early somite boundaries. Nature Genet. 19, 274–278 (1998).
Hrabe de Angelis, M., McIntyre, J. N. & Gossler, A. Maintenance of somite borders in mice requires the Delta homologue DII1. Nature 386, 717– 721 (1997).
Conlon, R. A., Reaume, A. G. & Rossant, J. Notch1 is required for the coordinate segmentation of somites. Development 121, 1533– 1545 (1995).
Swiatek, P. J., Lindsell, C. E., del Amo, F. F., Weinmaster, G. & Gridley, T. Notch1 is essential for postimplantation development in mice. Genes Dev. 8, 707– 719 (1994).
Heitzler, P. & Simpson, P. Altered epidermal growth factor-like sequences provide evidence for a role of Notch as a receptor in cell fate decisions. Development 117, 1113– 1123 (1993).
Klueg, K. M. & Muskavitch, M. A. Ligand-receptor interactions and trans-endocytosis of Delta, Serrate and Notch: members of the Notch signalling pathway in Drosophila. J. Cell Sci. 112, 3289–3297 (1999).
De Strooper, B. et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–522 (1999).
Schroeter, E., Kisslinger, J. & Kopan, R. Notch1 signalling requires ligand-induced proteolytic release of the intracellular domain. Nature 393, 382–386 (1998).
Brou, C. et al. A novel proteolytic cleavage involved in Notch signalling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5, 207–216 (2000).
Mumm, J. S. et al. A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol. Cell 5, 197–206 (2000).
MacArthur, C. A. et al. FGF-8 isoforms activate receptor splice forms that are expressed in mesenchymal regions of mouse development. Development 121, 3603-3613 (1995).
Hsieh, J. J. et al. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA 2. Mol. Cell. Biol. 16, 952-959 ( 1996).
Acknowledgements
We thank K. Irvine, A. Van der Bliek, E. Robey and G. Waters for critical and insightful comments on this work and K. Irvine, R. Haltiwanger and P. Stanley for communicating data before publication. We are also indebted to S. Ting-Berreth, T. Covey, C. MacArthur, G. Waters, D. Hayward, and J. Flanagan for reagents and J. Goodhouse for assistance with confocal imaging. C.H. and G.dS. were supported by NIH Training Grants awarded to UCLA and the MSTP, respectively, and S.H.J. was supported as a Harold W. Dodds Fellow of the Princeton University Graduate programme. This work was supported by grants from the NIH (NS 31885-05) (G.W.) and the Stop Cancer Foundation (G.W.) and the NIH (HD 30707) (T.F.V.) and funds provided to the Department of Molecular Biology by the Rathmann Family Foundation.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Hicks, C., Johnston, S., diSibio, G. et al. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat Cell Biol 2, 515–520 (2000). https://doi.org/10.1038/35019553
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/35019553
This article is cited by
-
Principles of human and mouse nephron development
Nature Reviews Nephrology (2022)
-
CCN1 interacts with integrins to regulate intestinal stem cell proliferation and differentiation
Nature Communications (2022)
-
Complex crosstalk of Notch and Hedgehog signalling during the development of the central nervous system
Cellular and Molecular Life Sciences (2021)
-
miR155 regulation of behavior, neuropathology, and cortical transcriptomics in Alzheimer's disease
Acta Neuropathologica (2020)
-
Impulsive control of a nonlinear dynamical network and its application to biological networks
Journal of Biological Physics (2019)