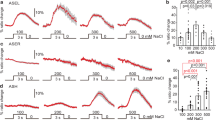
Abstract
The ability to discriminate between different chemical stimuli is crucial for food detection, spatial orientation and other adaptive behaviours in animals. In the nematode Caenorhabditis elegans, spatial orientation in gradients of soluble chemoattractants (chemotaxis) is controlled mainly by a single pair of chemosensory neurons1. These two neurons, ASEL and ASER, are left–right homologues in terms of the disposition of their somata and processes, morphology of specialized sensory endings, synaptic partners and expression profile of many genes2,3. However, recent gene-expression studies have revealed unexpected asymmetries between ASEL and ASER. ASEL expresses the putative receptor guanylyl cyclase genes gcy-6 and gcy-7, whereas ASER expresses gcy-5 (ref. 4). In addition, only ASEL expresses the homeobox gene lim-6, an orthologue of the human LMX1 subfamily of homeobox genes5. Here we show, using laser ablation of neurons and whole-cell patch-clamp electrophysiology, that the asymmetries between ASEL and ASER extend to the functional level. ASEL is primarily sensitive to sodium, whereas ASER is primarily sensitive to chloride and potassium. Furthermore, we find that lim-6 is required for this functional asymmetry and for the ability to distinguish sodium from chloride. Thus, a homeobox gene increases the representational capacity of the nervous system by establishing asymmetric functions in a bilaterally symmetrical neuron pair.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
01 August 2001
An Erratum to this paper has been published: https://doi.org/10.1038/35087630
References
Bargmann, C. I. & Horvitz, H. R. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7, 729–742 (1991).
Ward, S., Thompson, N., White, J. & Brenner, S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J. Comp. Neurol. 160, 313–338 (1975).
White, J., Southgate, E., Thompson, J. & Brenner, S. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil. Trans. R. Soc. Lond. B 314, 1–340 (1986).
Yu, S., Avery, L., Baude, E. & Garbers, D. L. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc. Natl Acad. Sci. USA 94, 3384–3387 (1997).
Hobert, O., Tessmar, K. & Ruvkun, G. The Caenorhabditis elegans lim-6 LIM homeobox gene regulates neurite outgrowth and function of particular GABAergic neurons. Development 126, 1547–1562 (1999).
Pierce-Shimomura, J. T., Morse, T. M. & Lockery, S. R. The fundamental role of pirouettes in Caenorhabditis elegans chemotaxis. J. Neurosci. 19, 9557–9569 (1999).
Ward, S. Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proc. Natl Acad. Sci. USA 70, 817–821 (1973).
Goodman, M. B., Hall, D. H., Avery, L. & Lockery, S. R. Active currents regulate sensitivity and dynamic range in C. elegans neurons. Neuron 20, 763–772 (1998).
Troemel, E. R., Sagasti, A. & Bargmann, C. I. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99, 387–398 (1999).
Bargmann, C. I., Thomas, J. H. & Horvitz, H. R. Chemosensory cell function in the behavior and development of Caenorhabditis elegans. Cold Spring Harb. Symp. Quant. Biol. 55, 529–538 (1990).
Bargmann, C. I. & Horvitz, H. R. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 251, 1243–1246 (1991).
Kaplan, J. M. & Horvitz, H. R. A dual mechanosensory and chemosensory neuron in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 90, 2227–2231 (1993).
Dusenbery, D. B. Analysis of chemotaxis in the nematode by counter current separation. J. Exp. Zool. 188, 41–47 (1974).
Bargmann, C. I., Hartwieg, E. & Horvitz, H. R. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74, 515–527 (1993).
Wes, P. D. & Bargmann, C. I. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature 410, 698–701 (2001).
Mori, K. & Yoshihara, Y. Molecular recognition and olfactory processing in the mammalian olfactory system. Prog. Neurobiol. 45, 585–619 (1995).
Rubin, B. D. & Katz, L. C. Optical imaging of odorant representations in the mammalian olfactory bulb. Neuron 23, 499–511 (1991).
Bargmann, C. I. & Avery, L. Laser killing of cells in Caenorhabditis elegans. Methods Cell Biol. 48, 225–250 (1995).
Sengupta, P., Chou, J. H. & Bargmann, C. I. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell 84, 899–909 (1996).
Lockery, S. R. & Goodman, M. B. Tight-seal whole-cell patch clamping of C. elegans neurons. Methods Enzymol. 295, 201–217 (1998).
Acknowledgements
We thank C. Bargmann and P. Wes for discussions and sharing unpublished data; O. Hobert for suggesting study of the lim-6 mutant; J. H. Thomas for technical instruction; O. Hobert, D. Garbers and The C. elegans Genetics Center for strains; M. Moravec and J. Cervantes for technical assistance; and J. Eisen, M. Goodman, T. Morse and M. Westerfield for discussion. This work was supported by the National Science Foundation; the National Institute of Mental Health; the National Heart, Lung, and Blood Institute; the Office of Naval Research; The Sloan Foundation; and The Searle Scholars Program.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Pierce-Shimomura, J., Faumont, S., Gaston, M. et al. The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature 410, 694–698 (2001). https://doi.org/10.1038/35070575
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35070575
This article is cited by
-
Plasticity in gustatory and nociceptive neurons controls decision making in C. elegans salt navigation
Communications Biology (2021)
-
Molecular and cellular determinants of motor asymmetry in zebrafish
Nature Communications (2020)
-
GABAergic motor neurons bias locomotor decision-making in C. elegans
Nature Communications (2020)
-
Rotatable microfluidic device for simultaneous study of bilateral chemosensory neurons in Caenorhabditis elegans
Microfluidics and Nanofluidics (2020)
-
Whole-animal connectomes of both Caenorhabditis elegans sexes
Nature (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.