Abstract
Cellular FLICE-inhibitory protein (c-FLIP) is an inhibitor of caspase-8 and is required for macrophage survival. Recent studies have revealed a selective role of caspase-8 in noncanonical IL-1β production that is independent of caspase-1 or inflammasome. Here we demonstrated that c-FLIPL is an unexpected contributor to canonical inflammasome activation for the generation of caspase-1 and active IL-1β. Hemizygotic deletion of c-FLIP impaired ATP- and monosodium uric acid (MSU)-induced IL-1β production in macrophages primed through Toll-like receptors (TLRs). Decreased IL-1β expression was attributed to a reduced activation of caspase-1 in c-FLIP hemizygotic cells. In contrast, the production of TNF-α was not affected by downregulation in c-FLIP. c-FLIPL interacted with NLRP3 or procaspase-1. c-FLIP is required for the full NLRP3 inflammasome assembly and NLRP3 mitochondrial localization, and c-FLIP is associated with NLRP3 inflammasome. c-FLIP downregulation also reduced AIM2 inflammasome activation. In contrast, c-FLIP inhibited SMAC mimetic-, FasL-, or Dectin-1-induced IL-1β generation that is caspase-8-mediated. Our results demonstrate a prominent role of c-FLIPL in the optimal activation of the NLRP3 and AIM2 inflammasomes, and suggest that c-FLIP could be a valid target for treatment of inflammatory diseases caused by over-activation of inflammasomes.
Similar content being viewed by others
Main
Death receptors initiate apoptotic signals through the formation of death-inducing signaling complexes (DISCs) containing FADD and procaspase-8. Cellular FLICE-inhibitory protein (c-FLIP) inhibits death receptor-induced apoptosis by blocking the activation of caspase-8 at DISC.1, 2, 3, 4, 5 c-FLIP is expressed in 3 isoforms: long form (c-FLIPL), short form (c-FLIPS), and Raji form (c-FLIPR). All three c-FLIP isoforms contain two death effector domains (DEDs), and their competition with procaspase-8 for FADD binding inhibits caspase-8 activation. c-FLIPL possesses an additional caspase-like domain (CLD) at its C terminus and, when present at low levels, may also promote caspase-8 activation. c-FLIP deficiency profoundly increases sensitivity to death receptor-mediated apoptosis.6, 7, 8 In addition to its prominent anti-apoptotic activity, c-FLIP is essential for embryonic development and T-cell development.6, 8 c-FLIP is also required for the development of macrophages.9
The inflammasome is assembled in macrophages and monocytes by inflammatory stimuli to process procaspase-1 into active caspase-1 and to generate mature IL-1β and IL-18.10, 11, 12, 13, 14 Different types of inflammasomes are activated in response to specific ligands or pathogens. For the AIM2 inflammasome, its activation is initiated by the binding of cytosolic dsDNA to AIM2. For NLRP3 inflammasomes, their assembly is activated by a wide variety of ligands such as microbes, microbial products, toxins, or aggregated particles. The formation of the NLRP3 inflammasome involves two sequential stages. In the first stage, signals from TLRs or other innate receptors activate NF-κB, leading to enhanced expression of pro-IL-1β and NLRP3.15 In the second stage, a distinct signal stimulates the binding of NLRP3 to ASC and procaspase-1. Cytosolic K+ reduction, reactive oxygen species, and lysosomal protease cathepsin B have been suggested as potential second signals for NLRP3 inflammasome formation, but the exact biochemical processes involved in inflammasome assembly remain incompletely understood.10, 11, 12, 14
Recent studies reveal that caspase-8 regulates specific inflammasome activation and IL-1β generation. Ligation of TLR3 and TLR4 has been shown to promote the processing of pro-IL-1β by caspase-8, independent of caspase-1 and NLRP3.16 Engagement of Dectin-1 induces activation of the noncanonical caspase-8 inflammasome, leading to caspase-1-independent generation of active IL-1β.17 Depletion of inhibitors of apoptosis protein (IAPs) by a SMAC mimetic in TLR-stimulated myeloid cells triggers the cleavage of IL-1β by caspase-8, in addition to activation of the canonical NLRP3-caspase-1 pathway.18 Stimulation of Fas also generates IL-1β in TLR-primed myeloid cells by the noncanonical caspase-8 pathway that is independent of ASC and caspase-1.19 In contrast, caspase-8 has been shown to inhibit NLRP3 inflammasome activation in the epidermis.20 A more recent study illustrates that caspase-8 does not affect conventional two-signal-induced NLRP3 inflammasome assembly, but suppresses NLRP3 inflammasome activated by LPS alone through blocking RIPK3-dependent processes.21 In the present study, we demonstrated that c-FLIP is required for optimal canonical NLRP3 and AIM2 inflammasome activation. Downregulation of c-FLIP impaired caspase-1 activation and IL-1β production. We found a specific interaction between c-FLIPL and NLRP3/procaspase-1/AIM2. In contrast, c-FLIP suppressed SMAC mimetic-induced and caspase-8-dependent IL-1β generation. Our results indicate a novel function of c-FLIP by which c-FLIP promotes the assembly of NLRP3 and AIM2 inflammasomes, and suggest that c-FLIP could be a target for treating auto-inflammatory diseases.
Results
c-FLIP knockdown decreases IL-1β production in THP-1 cells
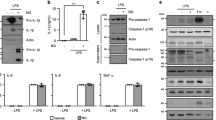
Using c-FLIP-specific shRNA, c-FLIP was downregulated in THP-1 cells, as shown by diminished levels of c-FLIPL and c-FLIPS (Figure 1a). The expression of NLRP3, procaspase-1, and ASC in c-FLIP-knocked down THP-1 cells was comparable to control cells (Figure 1a), while leaving the viability of THP-1 cells unaffected (Supplementary Figure 1). Treatment of PMA-primed THP-1 cells with NLRP3 inflammasome activators including R837, monosodium urate (MSU), R848, Pam3CSK4, or heat-killed Listeria monocytogenes (HKLM) led to IL-1β secretion (Figure 1b). The generation of IL-1β was attributed to the appearance of the cleaved caspase-1 and p17 IL-1β in cell supernatant (SUP) from stimulated THP-1 cells (Figures 1d–f). Pro-IL-1β was present in total-cell lysates (TCL) from PMA-primed THP-1 cells, and was further upregulated by treatment with Pam3CSK4 or HKLM, but not MSU (Figures 1d–f). The failure of MSU, alum crystals, or nigericin to upregulate pro-IL-1β in THP-1 cells is likely due to their inability to activate NF-κB (data not shown). Notably, Pam3CSK4 and HKLM also stimulated the expression of c-FLIP proteins (Figures 1e and f), which could be attributed to an induction of Cflar transcript by TLR (Supplementary Figure 2a). c-FLIP knockdown significantly reduced the secretion of IL-1β (Figure 1b). Pro-IL-1β induction was comparable between control and c-FLIP-deficient THP-1 cells stimulated by Pam3Csk4 or HKLM (Figures 1e and f). In contrast, the decrease in IL-1β production was associated with a diminished activation of caspase-1 following these treatments (Figures 1d–f), suggesting an impaired NLRP3 inflammasome function. As another control, TNF-α production was nearly identical between control and c-FLIP-knocked down THP-1 cells induced by R848, Pam3CSK4, or HKLM (Figure 1c). These results suggest that c-FLIP is required for the full activation of the NLRP3 inflammasome in THP-1 cells.
Attenuated IL-1β secretion in c-FLIP-knocked down THP-1 cells. (a) Downregulation of c-FLIP by shRNA in THP-1 cells. THP-1 cells were infected with pLL3.7 lentivirus (shCtr) or pLL3.7 containing c-FLIP-specific shRNA (shFLIP). The infected cells were sorted based on GFP expression, and the contents of c-FLIPL, c-FLIPR, NLRP3, procaspase-1 and ASC determined by immunoblots. Numbers to the left indicate molecular weight in kDa. (b and c) Reduced IL-1β secretion, but not TNF-α, in c-FLIP-deficient THP-1 cells. Control and c-FLIP-knocked down THP-1 cells were treated with PMA (500 ng/ml) for 3 h, cultured overnight, and then stimulated with R837 (10 μg/ml), MSU (150 μg/ml), R848 (2 μg/ml), Pam3CSK4 (Pam, 0.5 μg/ml), or HKLM (3 × 108/ml) 5 h. The secreted IL-1β (b) and TNF-α (c) in supernatants were determined by ELISA. (d–f) c-FLIP deficiency decreased procaspase-1 processing and IL-1β generation. Cell supernatants in (b) were precipitated with cold (−20 °C) acetone. Cells from (b) were also harvested and cell lysates prepared. Supernatant precipitates (SUP) and total cell lysates (TCL) were resolved on SDS-PAGE, and the expression of p17 IL-1β, procaspase-1, active p10 caspase-1, c-FLIPL, and pro-IL-1β determined. Error bars indicates S.D. of a specific experiment. **P<0.01, ***P<0.001 for paired t-test. All experiments (b–f) were repeated three times with similar results
c-FLIP hemizygotic deficiency impairs NLRP3 inflammasome activation in normal macrophages and dendritic cells
We next examined the effect of c-FLIP-knockout on the activation of NLRP3 inflammasome in macrophages. As previously reported, c-FLIP-knockout led to defective survival of macrophages,9, 22 and thioglycollate-elicited peritoneal macrophages were absent in c-FLIPf/f LysMCre mice (data not shown). Bone marrow cells from c-FLIPf/f LysMCre mice differentiated into macrophages after 6-day culturing in vitro were in numbers less than the control (c-FLIPf/f) bone marrow-derived macrophages (BMDMs). Consistent with previous analysis that the floxed Cflar allele is incompletely deleted in the surviving macrophages,9, 22 genomic DNA PCR revealed that c-FLIPf/f LysMCre BMDMs obtained were hemizygous (c-FLIP+/−) (Figure 2a). The reduction of c-FLIPL and c-FLIPR proteins in differentiated c-FLIPf/f LysMCre BMDMs was confirmed by immunoblots (Figure 2a). c-FLIPf/f LysMCre BMDMs used in the present study therefore were c-FLIP+/−, but not c-FLIP−/−, macrophages. In a separate experiment, we also generated c-FLIPf/− mice, in which all macrophages were hemizygous for the c-FLIP allele (Supplementary Figure 3a). The expression of NLRP3, ASC, and procaspase-1 was comparable between normal littermate (WT) and c-FLIPf/f LysMCre BMDMs (Figure 2a). Sequential treatment of BMDMs with LPS, R848, or Pam3CSK, followed by ATP, MSU, or nigercin led to secretion of IL-1β, whereas c-FLIP-hemi-deficiency significantly reduced the generation of IL-1β (Figures 2b and c, Supplementary Figure 3b). We first examined whether decreased IL-1β production was due to reduced cell survival in c-FLIP+/− BMDMs. Supplementary Figure 4 demonstrates that cell viability was not compromised in c-FLIP hemizygous macrophages before or after NLRP3 inflammasome stimulation. In addition, similar amounts of TNF-α were produced by control and c-FLIP+/− macrophages activated by LPS, R848, and Pam3CSK (Figure 2d, Supplementary Figure 3d). Furthermore, the induction of pro-IL-1β and NLRP3 proteins was comparable between control and c-FLIP hemizygotic BMDMs treated with LPS (Figures 2e and f, Supplementary Figures 3e and f). In addition, LPS-stimulated similar levels of Il1b and Nlrp3 transcripts in control and Cflar-deficient macrophages (Supplementary Figures 2b and c). We also evaluated the activation of NF-κB, essential for the expression of Il1b and Nlrp3, in Cflar+/− macrophages. LPS-induced RelA (p65) nuclear translocation was nearly identical between control and c-FLIP+/− macrophages (Supplementary Figure 5). Therefore, c-FLIP downregulation does not affect LPS-stimulated expression of pro-IL-1β and NLRP3. Instead, reduced IL-1β secretion in c-FLIP hemizygous macrophages was associated with a clear decrease in caspase-1 generation (Figures 2e and f, Supplementary Figures 3e and f), suggesting an attenuated activation of NLRP3 inflammasome in c-FLIP+/− macrophages.
Hemizygotic deletion of c-FLIP impairs caspase-1 activation and IL-1β generation in BMDMs. (a) Reduced c-FLIP expression in mouse BMDMs from c-FLIP hemizygotic mice. Bone marrow cells from FLIPf/f and FLIPf/f LysMCre mice were allowed to differentiate into macrophages. Analysis of c-FLIP floxed allele (FILPf) and deleted allele (FILPd) by PCR on genomic DNA from FLIPf/f and FLIPf/f LysMCre BMDMs indicated that FLIPf/f LysMCre BMDMs were hemizygous (FLIP+/−). The protein levels of c-FLIP, NLRP3, procaspase-1, and ASC were determined by western blot. (b–d) IL-1β production, but not TNF-α secretion, was reduced in c-FLIP+/− macrophages. BMDMs were primed with LPS (0.2 μg/ml) (b,d), R848 (2 μg/ml), or Pam3CSK4 (2 μg/ml) (c,d) for 4 h, and the secretion of TNF-α in the supernatants was determined (d). Cells were then activated by ATP (4 mM) for 45 min (b,c), or MSU (150 μg/ml) for 6 h (b). The secreted IL-1β in supernatants was determined. (e,f) Reduced caspase-1 activation in c-FLIP+/− BMDMs. Supernatant precipitate and total cell lysates from LPS/ATP (e)- or LPS/MSU (f)-stimulated BMDMs were prepared. The contents of p17 IL-1β, procaspase-1, active p10 caspase-1, NLRP3, pro-IL-1β and ASC were determined. Each data point (b–d) represents mean of triplicate determinations in a single experiment, and each experiment (a–f) was repeated three times with similar results. ***P<0.001 for paired t-test
We also examined the effect of c-FLIP deficiency in bone marrow-derived dendritic cells (BMDCs). The production of IL-1β stimulated by LPS/ATP or LPS/nigercin was attenuated in c-FLIPf/− BMDCs (Supplementary Figure 6b). The induction of pro-IL-1β was comparable between c-FLIPf/f and c-FLIPf/− BMDCs (Supplementary Figure 6c). A clear reduction in the generation of active p10 caspase-1 and mature p17 IL-1β was observed in c-FLIPf/− BMDCs (Supplementary Figure 6c). In contrast, the production of TNF-α in BMDCs stimulated by LPS was not altered by hemideficiency of c-FLIP (Supplementary Figure 6d).
Overexpression of c-FLIPL increases IL-1β generation
We next examined whether c-FLIP overexpression enhanced IL-1β generation in THP-1 cells. Transduction of c-FLIPL or c-FLIPS in THP-1 cells did not alter the amount of NLRP3, procaspase-1, or ASC (Figure 3a). We observed that alum crystal-, nigericin-, or MSU-induced secretion of IL-1β was increased in c-FLIPL-overexpressing THP-1 cells (Figure 3b). The cleavage of caspase-1 was also increased in c-FLIPL-transduced THP-1 cells (Figures 3d–f). Notably, c-FLIPL overexpression did not lead to processing of procaspase-8 in THP-1 cells (Supplementary Figure 7). In contrast to c-FLIPL, the expression of c-FLIPS (Figure 3a) failed to increase MSU- and nigericin-stimulated IL-1β generation (Figure 3c), indicating a specific stimulatory role of c-FLIPL in NLRP3 inflammasome activation.
Overexpression of c-FLIPL increases caspase-1 activation and IL-1β generation. (a) Overexpression of c-FLIP in THP-1 cells. THP-1 cells were infected with pTRIP, pTRIP-c-FLIPL-Myc, or pTRIP-c-FLIPS-Myc, and GFP-expressing cells were isolated by sorting. The expression of c-FLIP, NLRP3, caspase-1, and ASC were analyzed by immunoblots. (b) Increased IL-1β secretion in c-FLIPL-overexpressing THP-1 cells. PMA-primed THP-1 cells were stimulated with alum crystal (0.5 mg/ml), nigericin (10 μM), or MSU (150 μg/ml) for 6 h. The secreted IL-1β in the supernatants was determined. (c) c-FLIPS does not increase IL-1β secretion. THP-1 cells expressing vector, c-FLIPL, and c-FLIPS were compared for IL-1β production in response to MSU or nigericin stimulation. (d–f) Increased caspase-1 activation in c-FLIPL-overexpressing THP-1 cells. Supernatant precipitates (SUP) and total cell lysates (TCL) from (b) were analyzed for expression of p17 IL-1β, procaspase-1, active p10 caspase-1, and pro-IL-1β. Each data point represents mean of triplicate in a single experiment (b and c), and each experiment (a–f) was repeated three times with similar results. ***P<0.001 for paired t-test
c-FLIP is required for the activity of the reconstituted NLRP3 inflammasome
We also studied the requirement of c-FLIP for NLRP3 inflammasome reconstituted in 293T cells. c-FLIP was effectively knocked down in 293T cells by specific shRNA (Figure 4a). 293T cells do not express procaspase-1, NLRP3, or ASC, and introduction of these components could be readily detected (Figure 4b). Mature IL-1β was only generated in 293T cells with simultaneous reconstitution of pro-IL-1β, ASC, procaspase-1, and NLRP3 as measured by immunoblots and ELISA (Figures 4b and c). Knockdown of c-FLIP reduced the secretion of p17 IL-1β protein in 293T cells (Figures 4b and c). Therefore, c-FLIP is required for the activation of NLRP3 inflammasome reconstituted in 293T cells.
c-FLIP is required for functional reconstitution of NLRP3 inflammasome in 293T cells. (a) Knockdown of c-FLIP in 293T cells. 293T cells were infected with pLL3.7 (shCtr) or pLL-shFLIP (shFLIP), and GFP-positive cells were sorted. The expression of c-FLIPL was determined. (b and c) Decreased IL-1β secretion and reduced caspase-1 activation in c-FLIP-knocked down 293T cells. Control or c-FLIP-knocked down 293T cells were transfected with pro-IL-1β, NLRP3-myc, procaspase-1-myc, and ASC. The secretion of IL-1β in the supernatants 48 h after transfection was analyzed by ELISA (b). The contents of p17 IL-1β, NLRP3, pro-IL-1β, and ASC in supernatant precipitates (SUP) and total-cell lysates (TCL) were analyzed (c). Each data point represents triplicate determinations in a single experiment, and each experiment was repeated three times with similar results. ***P<0.001 for paired t-test
Interaction of c-FLIP with NLRP3 and procaspase-1
As the enhancing effect of c-FLIP on IL-1 production was mainly on caspase-1 processing, we examined the potential interaction of c-FLIP with components of the inflammasomes. We transfected 293T cells with NLRP3 and c-FLIPL, immunoprecipitated c-FLIPL, and found that NLRP3 was co-precipitated (Figure 5a). We also identified that the CLD of c-FLIPL interacted with NLRP3, whereas c-FLIPS did not bind NLRP3. Similarly, immunoprecipitation of full-length NLRP3 pulled down c-FLIPL (Figure 5b), which was likely mediated by the leucine-rich repeat (LRR) of NLRP3 (Figure 5c). We also detected an interaction of c-FLIPL with procaspase-1. Procaspase-1-HA was pulled down by FLAG-c-FLIPL or FLAG-CLD, but not c-FLIPS, when co-expressed in 293T cells (Figure 5d), indicating that CLD is the caspase-1-binding domain in c-FLIPL. In reciprocal precipitation, c-FLIPL was brought down by procaspase-1 and by the catalytic domain of procaspase-1, but not by the CARD domain of procaspase-1 (Figure 5e). No interaction between c-FLIPL and ASC, another element of the inflammasome, was detected in the same assay (Supplementary Figure 8).
Interaction of c-FLIPL with NLRP3 and procaspase-1. (a and b) Interaction between the CLD of c-FLIPL and NLRP3 in 293T cells. 293T cells were transfected with FLAG-c-FLIPL, FLAG-c-FLIPS, FLAG-CLD, and NLRP3-myc as indicated, and cell lysates were prepared 48 h later. The expression of the transfected proteins (Lysate) was examined by anti-Myc or anti-Flag. Cell lysates were precipitated with anti-FLAG (a) or anti-Myc (b) and analyzed by immunoblotting with anti-FLAG or anti-Myc. (c) The LRR domain of NLRP3 binds c-FLIPL. 293T cells were transfected with c-FLIPL-Myc, Flag-PYD, and Flag-LRR as indicated. Cell lysates were precipitated with anti-Flag, and the presence of c-FLIPL and NLRP3 fragments in precipitates and lysates determined. (d) The CLD of c-FLIP interacts with procaspase-1. 293T cells were transfected with Flag-c-FLIPL, FLAG-c-FLIPS, FLAG-CLD, and procaspase-1-HA as indicated, and cell lysates were precipitated with anti-Flag; the presence of procaspase-1 was detected by anti-HA. (e) The catalytic domain of procaspase-1 binds c-FLIP. 293T cells were transfected with procaspase-1-HA, procaspase-1(CARD)-HA, procaspase-1(Catalytic)-HA, and/or Flag-c-FLIPL, and cell lysates were prepared. The contents of c-FLIPL, procaspase-1 and its mutants in anti-HA precipitates and in cell lysates were determined
c-FLIP participates in formation of pyroptosome and inflammasome
We additionally used an ASC oligomerization assay to analyze the participation of c-FLIP in the function of the inflammasome. Caspase-1 activation has been linked to the formation of ASC pyroptosome, which contains ASC oligomers.23, 24 Supplementary Figure 9a illustrates the presence of ASC dimer, trimer, and tetramer in THP-1 cells stimulated with crude LPS and cell extracts treated with crosslinker disuccinimidyl suberate. Knockdown of c-FLIP decreased the extent of ASC oligomerization. Similarly, Pam3CSK4-stimulated ASC oligomerization was largely attenuated in c-FLIP-deficient THP-1 cells (Supplementary Figure 9b). A similar role of c-FLIP was found in ASC speck formation,24 another indicator of pyroptosome formation. In control THP-1 cells, treatment with nigericin led to a significant formation of ASC specks (Supplementary Figure 10). c-FLIP knockdown impaired the formation of ASC specks in THP-1. Therefore, c-FLIP deficiency prevents ASC oligomerization and decreases pyroptosome formation.
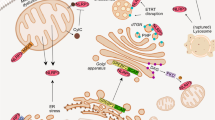
We also used in situ proximity ligation assay25 to measure the formation of inflammasomes in macrophages. The association of NLRP3 and procaspase-1 was undetectable in wild-type macrophages before and after LPS stimulation (Figure 6a), and the NLRP3-procaspase-1 interaction was visible only after adding ATP to LPS-primed macrophages (Figure 6a). A similar colocalization between NLRP3 and ASC was observed in macrophages treated with both LPS and ATP (Figure 6b). c-FLIP hemizygotic deficiency profoundly decreased the NLRP3-procaspase-1 association and NLRP3-ASC interaction in macrophages treated with LPS and ATP (Figures 6a and b). Recent studies have also revealed that inflammasome activation involves the mitochondrial localization of NLRP3. 26, 27 Figure 6c illustrates that nigercin promoted the colocalization of NLRP3 with Mitotracker,26 which was substantially decreased in c-FLIP-knocked down cells. These results suggest that c-FLIP is required for the full interaction of NLRP3-caspase-1 and NLRP3-ASC, as well as the situation of NLRP3 in mitochondria, during inflammasome assembly.
c-FLIP deficiency reduces in situ association of NLRP3, ASC, and caspase-1 and decreases NLRP3 mitochondrial localization. (a,b) Decreased NLRP3-caspase-1 association and reduced NLRP3-ASC interaction in activated c-FLIP-deficient macrophages. FLIPf/f or FLIPf/f LysMCre BMDMs were treated, as indicated, with LPS (0.2 μg/ml) for 4 h followed by ATP (2.5 mM) for 25 min. Cells were fixed by 4% paraformaldehyde and then stained with anti-NLRP3 and anti-caspase-1 (a), or anti-NLRP3 and anti-ASC (b). Oligonucleotide-conjugated secondary antibodies and the template oligonucleotides were sequentially added. The primers in close proximity were annealed, circularized, and amplified by rolling-circle amplification. The amplicons were detected by hybridization with a fluorescence probe, and analyzed by confocal microscopy. Bar indicates 10 μm. For quantification, individual amplicons were counted in four different fields by ImageJ and divided by the number of cells to obtain spots/nucleus. ***P<0.001 for paired t-test. (c) c-FLIP-knocked down decreases mitochondrial localization of NLRP3. 293T cells stably expressing NLRP3-HA were mock-transfected or transfected with siFLIP. Cells were untreated or treated with nigericin (10 μM) for 1 h. The confocal images were collected and quantitated by ZEN imaging software. Mito, Mitotracker Red. Coloc, colocalization (displayed in white spots). Bar indicates 20 μm
We further determined the association of c-FLIP with NLRP3 in macrophages after inflammasome formed. In situ proximity ligation assay revealed that close approximation of c-FLIP with NLRP3 was induced by the dual signals that promote inflammasome assembly (Supplementary Figure 11c). In a different analysis, the interaction between c-FLIP and NLRP3 could be detected by immunoprecipitation in BMDMs treated with LPS and nigericin (Supplementary Figure 11a). We further examined the association between NLRP3 and c-FLIP in inflammasome activation in vitro.28 Supplementary Figure 11b illustrates that mature IL-1β could be detected 15 min after incubation of BMDM lysates at 30 °C, accompanied by a clear association of c-FLIP with NLRP3 in immunoprecipitation assay. These results suggest that c-FLIP becomes associated with NLRP3 upon inflammasome formation.
c-FLIP is required for full AIM2 inflammasome activation
We further examined whether c-FLIP regulated the activation of other inflammasomes. The activation of AIM2 inflammasome is triggered by cytosolic dsDNA. Transfection of THP-1 cells with dAdT led to IL-1β production, which was significantly reduced in c-FLIP-knocked down THP-1 cells (Figure 7a). Overexpression of c-FLIPL enhanced AIM2 inflammasome-mediated IL-1β generation in THP-1 cells (Figure 7b). In addition, dsDNA-induced IL-1β production was attenuated in c-FLIPf/− BMDMs (Figure 7c, Supplementary Figure 3c). We also found that AIM2 interacted with c-FLIP. c-FLIPL or CLD, but not c-FLIPS, pulled down AIM2-HA when co-expressed in 293T cells (Figure 7d). Immunoprecipitation of AIM2 or AIM2(HIN), but not AIM2(PYD), also brought down c-FLIPL (Figure 7e). These results suggest that the CLD of c-FLIPL binds the HIN region of AIM2. Therefore, c-FLIPL interacts with AIM2 and participates in the activation of the AIM2 inflammasome.
c-FLIPL interacts with AIM2 and is required for full activation of AIM2 inflammasome. (a) AIM2-stimulated IL-1β production was decreased in c-FLIP-knockdown THP-1 cells. PMA-primed control (shCtr) and c-FLIP-knocked down (shFLIP) THP-1 cells were transfected with dAdT using Lipofectamine 2000, and production of IL-1β was determined 6 h later. (b) c-FLIPL increased the activation of AIM2 inflammasome in THP-1 cells. PMA-primed control (pTRIP) and c-FLIPL-transduced THP-1 cells were transfected with dAdT, and the secretion of IL-1β was determined 6 h later. (c) Reduced AIM2 inflammasome in c-FLIP+/− BMDMs. FLIPf/f or FLIPf/f LysMCre BMDMs were primed with LPS for 3 h, followed by alum or transfection with dAdT, and IL-1β secretion was quantitated 6 h later. Experiments (a–c) were independently repeated three times. ***P<0.001 for paired t-test. (d and e) Interaction between CLD of c-FLIPL and HIN region of AIM2. 293T cells were transfected with c-FLIPL, c-FLIPS, CLD, AIM2, AIM2(PYD), or AIM2(HIN) as indicated. Cell lysates were immunoprecipitated with anti-Flag (d) or anti-HA (e). The levels of AIM2, AIM2 truncated mutants, and c-FLIP in precipitates and cell lysates were determined
c-FLIP hemizygotic deficiency increases SMAC mimetic-, FasL, or Dectin-1-induced IL-1β generation
Depletion of IAPs by genetic manipulation or a SMAC mimetic in TLR-primed macrophages triggers the cleavage of IL-1β by caspase-8, in addition to the canonical NLRP3-caspase-1 pathway.18 We also examined the role of c-FLIP in SMAC mimetic-induced IL-1β production, because c-FLIP specifically suppresses caspase-8 activity. In wild-type macrophages primed with LPS, the SMAC mimetic AT-406 induced modest IL-1β secretion (Figure 8a). A large portion of IL-1β processing induced by LPS and AT-406 was mediated by caspase-8, as shown by their sensitivity to z-IETD inhibition (Figure 8b). In contrast, IL-1β production stimulated by LPS plus ATP in BMDMs was not affected by z-IETD (data not shown), consistent with recent report that caspase-8 is not involved in conventional NLRP3 inflammasome activation.21 c-FLIP hemizygotic deletion enhanced the production of IL-1β (Figures 8a and b). Similar to a previous report,18 AT-406 treatment generated both caspase-1 and caspase-8 (Figure 8c). c-FLIP hemizyogotic deficiency increased the processing of procaspase-8, but did not affect the cleavage of procaspase-1. Consequently, more p17 IL-1β was generated in c-FLIP+/− macrophages activated by LPS and AT-406 (Figure 8c).
IAP inhibitor-induced IL-1β production is inhibited by c-FLIP. (a and b) SMAC mimetic AT-406-stimulated, caspase-8-dependent IL-1β secretion was increased in c-FLIP+/− BMDMs. FLIPf/f or FLIPf/f LysMCre BMDMs were primed with or without LPS for 3 h, followed by AT-406 (a) and/or Z-IETD (b) as indicated, and IL-1β production was determined 6 h later. (c) c-FLIP deficiency increased the processing of procaspase-8 but not the cleavage of procaspase-1. Supernatant precipitate and total cell lysates were prepared from BMDMs in (a). The contents of procaspase-1, caspase-1, and IL-1β in supernatants, and the levels of p43/41 caspase-8, p18 caspase-8, procaspase-1, pro-IL-1β, and cIAP1 in total cell lysates were determined. ***P<0.001 for paired t-test
We further determined the regulatory role of c-FLIP in FasL-induced and caspase-8-dependent IL-1β production.19 Priming with LPS led to comparable Fas expression in c-FLIPf/f and c-FLIPf/− BMDMs (Supplementary Figure 12a). Treatment with FasL generated IL-1β in BMDMs. FasL-induced IL-1β secretion was suppressed by the addition of z-IETD, illustrating the requirement of caspase-8 in IL-1β processing. Consistent with a specific inhibition of caspase-8 by c-FLIP, Fas-mediated IL-1β generation was increased in c-FLIP+/− BMDMs (Supplementary Figure 12b). We also examined Dectin-1-mediated and caspase-8-dependent IL-1β generation17 in c-FLIP hemizygotic macrophages. Heat-killed Candida albicans (HKCA)-induced IL-1β production was inhibited by z-IETD, confirming the involvement of caspase-8 (Supplementary Figure 12c). Dectin-1-mediated IL-1β secretion was enhanced by more than threefold in FLIPf/f LysMCre BMDMs, relative to FLIPf/f BMDMs. Therefore, c-FLIP has an inhibitory role in IAP inhibitor-, FasL-, or HKCA-triggered IL-1β secretion, in contrast to the stimulatory role of c-FLIP in the NLRP3 inflammasome and AIM2 inflammasome.
Discussion
The assembly of the inflammasome is essential for the processing of procaspase-1 and pro-IL-1β. With the exception of the NLRP1 inflammasome,29 the detailed biochemical processes in the assembly of the inflammasome remain largely unclear. In the present study we identify a novel role of c-FLIP in the full activation of NLRP3 and AIM2 inflammasome. c-FLIP hemizygotic deficiency impaired procaspase-1 cleavage and IL-1β maturation, whereas c-FLIP overexpression enhanced caspase-1 processing and IL-1β production (Figures 1, 2, 3, Supplementary Figures 3 and 6). We also demonstrated that the formation of inflammasome in situ was diminished in the absence of c-FLIP (Figure 6). c-FLIPL interacts with NLRP3 and procaspase-1 when co-expressed (Figure 5). In addition, we observed a close association of c-FLIP with NLRP3 during inflammasome assembly (Supplementary Figure 11).
c-FLIP is an inhibitor of death receptor-mediated apoptosis and necrosis.1, 30, 31 Complete deletion of c-FLIP leads to significant death of macrophages.9, 22 We measured cell death before or after inflammasome activation but did not find any increase in cell death in c-FLIP+/− macrophages (Supplementary Figure 1 and 4). In addition, we did not find that c-FLIP downregulation had any effect on the production of TNF-α (Figures 1 and 2, Supplementary Figures 3 and 6). Furthermore, hemizygous c-FLIP deficiency decreased NLRP3- and AIM2-mediated IL-1β generation (Figures 2 and 7), but increased AT-406- and FasL-triggered IL-1β secretion in BMDMs (Figure 8, Supplementary Figure 12). Therefore, the diminished activation of NLRP3 and AIM2 inflammasomes in c-FLIP-downregulated macrophages was not due to reduced survival of macrophages.
c-FLIP has been shown to activate NF-κB,32, 33 which is required for the transcription of pro-IL-1β and NLRP3, suggesting a possibility that the observed reduction in IL-1β secretion is due to attenuated expression of pro-IL-1β and NLRP3. We found normal NF-κB activation in c-FLIP+/− BMDMs (Supplementary Figure 5). During the activation of the inflammasome, the induction of pro-IL-1β and NLRP3 mRNA (Supplementary Figure 2), as well as the upregulation of pro-IL-1β and NLRP3 protein (Figures 2e and f, Supplementary Figure 3 and 6), were not affected by c-FLIP hemizygous deficiency. In addition, the expression of TNF-α, encoded by another NF-κB-dependent gene, was not affected by c-FLIP downregulation (Figure 2d, Supplementary Figure 3d and Figure 6d). Therefore, c-FLIP is not essential for TLR-induced NF-κB activation in macrophages.
We also found that c-FLIP is upregulated by inflammatory stimuli including LPS, Pam3CSK4, and HKLM in wild-type macrophages (Figure 1, Supplementary Figures 2a, 3e, and 3f). NF-κB activation represents a common signaling process downstream of various TLRs, while c-FLIP is a well-known target transactivated by NF-κB. Therefore, the first stage of inflammasome activation involves the induction of c-FLIP, in addition to the upregulation of pro-IL-1β and NLRP3. Our results thus suggest a possibility that that TLR-induced increase in c-FLIP expression ensures that there are sufficient amounts of c-FLIP to assist during the assembly of the inflammasome that will then promote the generation of active caspase-1.
A recent study revealed that caspase-8 deficiency leads to skin inflammation, caused by increased inflammasome formation and IL-1β production.20 c-FLIP, by itself a caspase-8 inhibitor, may exert its function through this inhibition. However, the enhanced inflammasome formation in caspase-8-null epidermal cells is attributed to increased expression of NLRP3,20 yet there was no change in NLRP3 levels in c-FLIP-overexpressing or c-FLIP-knocked down macrophages (Figures 1, 2, 3), suggesting a difference between epidermal cells and macrophages. In a more recent study, caspase-8 deficiency sensitized dendritic cells to produce IL-1β by stimulation with LPS alone, but caspase-8 is not involved in NLRP3 inflammasome activation induced by both LPS and ATP.21 The participation of c-FLIP in NLRP3 and AIM2 inflammasome activation reported in this study is thus unlikely mediated by inhibition of caspase-8 activity. In addition, we examined the role of c-FLIP in caspase-8-dependent inflammasome activation stimulated by SMAC mimetic, Dectin-1, or FasL,17, 18, 19 and found that c-FLIP suppressed IL-1β generation by inhibition of caspase-8 (Figure 8, Supplementary Figure 12), demonstrating opposing functions of c-FLIP in NLRP3/AIM2 and caspase-8-dependent inflammasome activation.
Our results illustrating that c-FLIP is required for the activation of both the NLRP3 inflammasome and AIM2 inflammasome suggest that c-FLIP may act at a stage common to the activation of different inflammasomes. c-FLIP inhibits autophagy,31, 34 and autophagy has been shown to inhibit inflammasome formation.35, 36, 37 Therefore, it is possible that the inhibition of autophagy by c-FLIP is required for the activation of the inflammasome. Notably, we found that c-FLIPL interacted with NLRP3/procaspase-1. c-FLIPS, the c-FLIP isoform that does not bind NLRP3/procaspase-1 (Figure 5), did not promote inflammasome activation (Figure 3). In addition, we identified that c-FLIPL is part of NLRP3 inflammasome formed in vivo and in vitro (Supplementary Figure 11). Future work will help identify the exact mechanism orchestrating c-FLIP-assisted inflammasome formation.
Aberrant NLRP3 inflammasome activation contributes to auto-inflammatory diseases as well as inflammasome-associated diseases including gout, silicosis, asbestosis, atherosclerosis, Alzheimer’s disease, and type 2 diabetes mellitus.38, 39 A number of anti-inflammatory molecules have been developed for treatment of inflammasome- and IL-1β-induced diseases.40 As c-FLIP is upregulated in many tumor cells and is an important target for cancer therapy, small molecule drugs that inhibit the expression of c-FLIP or induce the degradation of c-FLIP have been developed, in addition to c-FLIP-specific siRNA.41 The present studies suggest a possibility that c-FLIP-downregulating compounds may be used to counteract the over-activation of the inflammasome. Future experiments will help determine the feasibility of controlling inflammasome-associated diseases by targeting c-FLIP.
Materials and Methods
Reagents
Purified LPS, R848, HKLM, Pam3CSK4, nigericin, alum crystals, HKCA, and monosodium urate (MSU) were purchased from InvivoGen (San Diego, CA, USA). Anti-ASC (AL177), anti-NLRP3 (Cryo-2), anti-human FLIP (NF-6), and anti-mouse FLIP (Dave-2) antibodies were obtained from ENZO Life Sciences (Farmingdale, NY, USA). Anti-mouse caspase-1 (M20), anti-human caspase-1 (A19), anti-c-FLIP (H-150 and G-11) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-mature IL-1β (Asp116) and anti-Myc (9B11) antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). AT-406 was synthesized according to Cai et al.42 Crude LPS, PMA, Flag-M2 antibodies, and Flag-M2 beads were purchased from Sigma-Aldrich (St. Louis, MO, USA). For ELISA assays, human IL-1β DuoSet, human TNF-α DuoSet, murine IL-1β DuoSet, and murine TNF-α DuoSet were obtained from R&D (Minneapolis, MN, USA).
Mice
c-FLIPf/f mouse was previously described.8 c-FLIPf/f mice were bred with LysMCre mice43 (The Jackson Laboratory, Bar Harbor, ME, USA) to generate c-FLIPf/f LysMCre mice. Previous studies have shown that macrophage cannot survive with homozygous deletion of c-FLIP.22 Genomic PCR indicated that FLIPf/f LysMCre BMDMs are c-FLIP hemizygote (FLIP+/−), and are abbreviated as FLIP+/−. c-FLIPf/f mice were also crossed with B6 mice to produce c-FLIPf/+ mice. c-FLIPf/+ mice were bred with E2A-Cre mice, followed by back-crossing to c-FLIPf/f mice to select for c-FLIPf/− mice without E2A-Cre. PCR primers used for genomic DNA typing were FLIPSAF1, CATGAGCACTGAGGGACACAGCAC; FLIPFlox, CGGAGTTTGCTACAGGAAGGCCAC (generated 480 bp PCR product with FLIPSAF1); FLIPd, AGGCTAGTTAACTGGCTCAGC (generated 800 bp PCR product with FLIPSAF1). All mouse experiments were conducted with approval from the Institutional Animal Care and Utilization Committee, Academia Sinica.
Cell culture
Human monocyte cell line THP-1 was cultured in RPMI1640 medium supplemented with 10% fetal bovine serum (Life Technologies-Invitrogen, Carlsbad, CA, USA), 10 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 20 μM 2-ME (complete RPMI medium). Bone marrow cells were collected from tibias and femurs by flushing with cold PBS with a 25G needle. Bone marrow cells were cultured either in complete DMEM medium (same supplements as complete RPMI) with 20% L929 conditioned media for 6 days to generate BMDMs, or in DMEM containing 20 ng/ml GM-CSF (R&D) for 8 days to generate BMDCs.
c-FLIP overexpression and knockdown
For overexpression of c-FLIP, c-FLIP cDNA44, 45 was subcloned into pTRIP-IRES-GFP to generate pTRIP-c-FLIP-IRES-GFP. 293T cells were transfected with 20 μg pTRIP-IRES-GFP or pTRIP-c-FLIP-IRES-GFP, 15 μg psPAX2, and 6 μg pMD2G, and lentivirus-containing culture supernatants were harvested. THP-1 cells were infected with recombinant lentivirus, and GFP-expressing cells were isolated 48 h post-infection by sorting on a FACSVantage SE (BD Biosciences, San Jose, CA, USA). For c-FLIP knockdown, human c-FLIP-specific shRNA were subcloned into pLentiLox vector (pLL3.7; gift from Dr. Luk Van Parijs, Massachusetts Institute of Technology, Cambridge, MA, USA). The sequence of the human c-FLIP-specific shRNA was 5′-GCA GTC TGT TCA AGG AGC A. Lentiviruses were harvested from culture supernatants of 293T cells transfected with pLL3.7 or pLL3.7-shFLIP (20 μg), psPAX2 (15 μg), and pMD2.G (6 μg). THP-1 cells were infected with recombinant lentivirus, and GFP-expressing cells were sorted 48 h post-infection on a FACSVantage SE (Becton Dickinson, Mountain View, CA, USA). c-FLIP was also knocked down in THP-1 and 293T cells by transfection with siFILP (same sequence as shRNA) using Lipofectamine 2000.
Measurement of IL-1β
THP-1 cells were treated with 0.5 μg/ml phorbol myristate acetate (PMA) for 3 h, replenished with complete RPMI medium for overnight culture. Cells were activated by R837 (10 μg/ml), R848 (2 μg/ml), Pam3CSK4 (0.5 μg/ml), MSU (150 μg/ml), alum crystal (0.5 mg/ml), HKLM (3 × 108/ml), nigericin (10 μM), or crude LPS (15 μg/ml) in serum-free RPMI for 6 h. Both cells and culture supernatants were harvested. The contents of IL-1β or TNF-α supernatants were quantitated by ELISA. The remaining culture supernatants were precipitated by cold acetone (−20 °C). Both total cell lysates and supernatant precipitates were resolved by 4–20% gradient SDS-PAGE and analyzed for pro-IL-1β, IL-1β, NLRP3, procaspase-1, and caspase-1 by immunoblots. For BMDMs, cells were primed by purified LPS (0.2 μg/ml), R848 (2 μg/ml), or Pam3CSK4 (2 μg/ml) for 4 h, and the culture supernatants were collected and analyzed for TNF-α secretion. Cells were then activated by ATP (5 mM) or nigericin (10–20 μM) for 45 min or MSU (150 μg/ml) for 6 h. Both cells and culture supernatants were isolated, and analyzed by immunoblots as described for THP-1 cells.
Cell viability assay
Cell viability was determined by measuring ATP levels. LPS-primed BMDMs were treated with MSU or alum for 3 h. Equal volume of Cell Titer-Glo reagent (Promega, Madison, WI, USA) was then added and incubated for an additional 30 min. Luminescent signal was determined using a luminescence reader, the Victor3 1420 Multilabel Counter (PerkinElmer, Shelton, CT, USA).
NLRP3 inflammasome reconstitution
293T cells were infected with recombinant lentivirus pLL3.7 or pLL3.7-shFLIP, and GFP-expressing cells were sorted 48 h post-infection. Control and pLL-shFLIP 293T cells were then transfected with pcDNA4-pro-IL-1β, pcDNA4-NLRP3-Myc, pcDNA4-ASC, and pcDNA4-procaspase-1-Myc using Lipofectamine 2000 (Invitrogen). Both cells and the culture supernatants were collected 48 h later, and analyzed by ELISA and immunoblots as described for THP-1 and BMDMs.
Pyroptosome
THP-1 cells were treated with PMA for 3 h, washed and incubated overnight. Cells were stimulated with crude LPS (5 μg/ml) or Pam3Csk4 (1 μg/ml) for 2 h, harvested and lysed in buffer A (20 mM HEPES, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 320 mM sucrose). The bulk nuclei were removed by centrifugation, after which the supernatant was diluted with equal volume of buffer A and filtered. The filtrate was diluted with an equal volume of CHAPS buffer (20 mM HEPES (pH 7.5), 5 mM MgCl2, 0.5 mM EGTA, 0.1 mM PMSF, 0.1% CHAPS) and centrifuged; the resulting pellet contained the ASC pyroptosomes. The pellets were resuspended in CHAPS buffer and then subjected to crosslinking using disuccinimidyl suberate (DSS) (2 mM) for 30 min. The pyroptosomes were resolved on 10% SDS-PAGE, and the levels of ASC and caspase-1 analyzed. The formation of pyroptosomes was also analyzed in ASC-mCherry-expressing THP-1 cells according to published method.24 The ASC pyroptosomes images were acquired in Olympus (Tokyo, Japan) 1 × 71 fluorescence microscope and Zeiss (Oberkochen, Germany) LSM 510 confocal laser scanning.
In situ proximity ligation assay
In situ proximity ligation assay was performed according to Söderberg et al.25 using the Duolink Proximity Ligation in situ reagent kit (Olink, Uppsala, Sweden). The detailed procedures were recently described.46 The cells were examined under a Zeiss LSM 510 confocal laser scanning microscope with a × 63 objective lens.
NLRP3 mitochondrial localization
NLRP3 mitochondrial localization was performed as described.26 293T cells were stained with Mitotracker Red (Invitrogen) followed by permeabilization and fixation. Cells were then sequentially stained with anti-HA (for NLRP3-HA) and Alexa Fluro-conjugated anti-mouse. Images were acquired on a Zeiss LSM 510 confocal laser scanning microscope. The colocalization of NLRP3-HA and Mitotracker was quantitated using ZEN imaging software ZEN 2010 version 6.0. Each group contains at least 10 cells.
Quantitative PCR
Total RNA from control and c-FLIP-knockout BMDM cells was isolated using TRIzol (Invitrogen). cDNAs were prepared and analyzed for the expression of Cflar, Nlrp3, and Il1b on a LightCycler 480 Real-Time PCR System (Roche, Mannheim, Germany). The PCR protocol is 95 °C for 10 min, followed by 45 cycles of 95 °C for 10 s, 60 °C annealing for 10 s, and 72 °C extension for 8 s. The PCR primers were as follows: Cflar, forward, 5′-GGCAAGATAGCCAAGGACA-3′ and reverse, 5′-CGAAGCCTGGAGAGTATTCAT-3′; Nlrp3, forward, 5′-CGAGACCTCTGGGAAAAAGCT-3′ and reverse, 5′-GCATACCATAGAGGAATGTGATGTACA-3′; Il1b, forward, 5′-CGGCACACCCACCCTG-3′ and reverse, 5′-AAACCGCTTTTCCATCTTCTTCT-3′.
Abbreviations
- AIM2:
-
HIN-200 protein absent in melanoma 2
- ASC:
-
apoptosis-associated speck-like protein containing a CARD domain
- BMDC:
-
bone marrow-derived dendritic cell
- BMDM:
-
bone marrow-derived macrophage
- c-FLIP:
-
cellular FLICE-inhibitory protein
- CLD:
-
caspase-like domain
- HKCA:
-
heat-killed Candida albicans
- HKLM:
-
heat-killed Listeria monocytogenes
- IAP:
-
inhibitor of apoptosis proteins
- LRR:
-
leucine-rich repeat
- MSU:
-
monosodium uric acid
- NLRP3:
-
NACHT domain-, leucine-rich repeat-, and PYD-containing protein 3
- PYD:
-
pyrin domain
- SMAC:
-
second mitochondria-derived activator of caspases
- TLR:
-
Toll-like receptor
References
Budd RC, Yeh WC, Tschopp J . cFLIP regulation of lymphocyte activation and development. Nat Rev Immunol 2006; 6: 196–204.
Strasser A, Jost PJ, Nagata S . The many roles of FAS receptor signaling in the immune system. Immunity 2009; 30: 180–192.
Wilson NS, Dixit V, Ashkenazi A . Death receptor signal transducers: nodes of coordination in immune signaling networks. Nat Immunol 2009; 10: 348–355.
Lavrik IN, Krammer PH . Regulation of CD95/Fas signaling at the DISC. Cell Death Differ 2012; 19: 36–41.
Dickens LS, Powley IR, Hughes MA, MacFarlane M . The 'complexities' of life and death: death receptor signalling platforms. Exp Cell Res 2012; 318: 1269–1277.
Yeh WC, Itie A, Elia AJ, Ng M, Shu HB, Wakeham A et al. Requirement for Casper (c-FLIP) in regulation of death receptor-induced apoptosis and embryonic development. Immunity 2000; 12: 633–642.
Chau H, Wong V, Chen NJ, Huang HL, Lin WJ, Mirtsos C et al. Cellular FLICE-inhibitory protein is required for T cell survival and cycling. J Exp Med 2005; 202: 405–413.
Zhang N, He YW . An essential role for c-FLIP in the efficient development of mature T lymphocytes. J Exp Med 2005; 202: 395–404.
Huang QQ, Perlman H, Huang Z, Birkett R, Kan L, Agrawal H et al. FLIP: a novel regulator of macrophage differentiation and granulocyte homeostasis. Blood 2010; 116: 4968–4977.
Lamkanfi M, Dixit VM . Inflammasomes: guardians of cytosolic sanctity. Immunol Rev 2009; 227: 95–105.
Latz E . The inflammasomes: mechanisms of activation and function. Curr Opin Immunol 2010; 22: 28–33.
Davis BK, Wen H, Ting JP . The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol 2011; 29: 707–735.
Strowig T, Henao-Mejia J, Elinav E, Flavell R . Inflammasomes in health and disease. Nature 2012; 481: 278–286.
Rathinam VA, Vanaja SK, Fitzgerald KA . Regulation of inflammasome signaling. Nat Immunol 2012; 13: 333–342.
Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D et al. Cutting edge: NF-κB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol 2009; 183: 787–791.
Maelfait J, Vercammen E, Janssens S, Schotte P, Haegman M, Magez S et al. Stimulation of Toll-like receptor 3 and 4 induces interleukin-1beta maturation by caspase-8. J Exp Med 2008; 205: 1967–1973.
Gringhuis SI, Kaptein TM, Wevers BA, Theelen B, van der Vlist M, Boekhout T et al. Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1β via a noncanonical caspase-8 inflammasome. Nat Immunol 2012; 13: 246–254.
Vince JE, Wong WW, Gentle I, Lawlor KE, Allam R, O’Reilly L et al. Inhibitor of apoptosis proteins limit RIP3 kinase-dependent interleukin-1 activation. Immunity 2012; 36: 215–227.
Bossaller L, Chiang PI, Schmidt-Lauber C, Ganesan S, Kaiser WJ, Rathinam VA et al. Cutting Edge: FAS (CD95) mediates noncanonical IL-1β and IL-18 maturation via caspase-8 in an RIP3-independent manner. J Immunol 2012; 189: 5508–5512.
Lee P, Lee DJ, Chan C, Chen SW, Ch’en I, Jamora C . Dynamic expression of epidermal caspase 8 simulates a wound healing response. Nature 2009; 458: 519–523.
Kang TB, Yang SH, Toth B, Kovalenko A, Wallach D . Caspase-8 blocks kinase RIPK3-mediated activation of the NLRP3 inflammasome. Immunity 2013; 38: 27–40.
Gordy C, Pua H, Sempowski GD, He YW . Regulation of steady-state neutrophil homeostasis by macrophages. Blood 2011; 117: 618–629.
Fernandes-Alnemri T, Alnemri ES . Assembly, purification and assay of the activity of the ASC pyroptosome. Methods Enzymol 2008; 442: 251–270.
Fernandes-Alnemri T, Wu J, Wu JW, Datta P, Miller B, Jankowski W et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Diff 2007; 14: 1590–1604.
Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius KJ, Jarvius J et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Method 2006; 3: 995–1000.
Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN . The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell 2013; 153: 348–361.
Misawa T, Takahama M, Kozaki T, Lee H, Zou J, Saitoh T et al. Microtubule-drivenspatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat Immunol 2013; 14: 454–460.
Martinon F, Burns K, Tschopp J . The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-1β . Mol Cell 2002; 10: 417–426.
Faustin B, Lartigue L, Bruey JM, Luciano F, Sergienko E, Bailly-Maitre B et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell 2007; 25: 713–724.
Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C et al. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 2011; 471: 363–367.
He MX, He YW . A role for c-FLIPL in the regulation of apoptosis, autophagy, and necroptosis in T lymphocytes. Cell Death Differ 2013; 20: 188–197.
Kataoka T, Budd RC, Holler N, Thome M, Martinon F, Irmler M et al. The caspase-8 inhibitor FLIP promotes activation of NF-κB and Erk signaling pathways. Curr Biol 2000; 10: 640–648.
Golks A, Brenner D, Krammer PH, Lavrik IN . The c-FLIP-NH2 terminus (p22-FLIP) induces NF-κB activation. J Exp Med 2006; 203: 1295–1305.
Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR et al. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol 2009; 11: 1355–1362.
Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 2008; 456: 264–268.
Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol 2011; 12: 222–230.
Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA et al. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol 2012; 13: 255–263.
Kastner DL, Aksentijevich I, Goldbach-Mansky R . Autoinflammatory disease reloaded: a clinical perspective. Cell 2010; 140: 784–790.
Rock KL, Latz E, Ontiveros F, Kono H . The sterile inflammatory response. Annu Rev Immunol 2010; 28: 321–342.
Dinarello CA . Anti-inflammatory agents: present and future. Cell 2010; 140: 935–950.
Safa AR, Pollok KE . Targeting the anti-apoptotic protein c-FLIP for cancer therapy. Cancers (Basel) 2011; 3: 1639–1671.
Cai Q, Sun H, Peng Y, Lu J, Nikolovska-Coleska Z, McEachern D et al. A potent and orally active antagonist (SM-406/AT-406) of multiple inhibitor of apoptosis proteins (IAPs) in clinical development for cancer treatment. J Med Chem 2011; 54: 2714–5426.
Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I . Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 1999; 8: 265–277.
Yeh JH, Hsu SC, Han SH, Lai MZ . Mitogen activated kinase kinase antagonized FADD-mediated apoptosis by induced FLIP expression. J Exp Med 1998; 188: 1795–1802.
Fang LW, Tai TS, Yu WN, Liao F, Lai MZ . Phosphatidylinositide 3-kinase priming couples c-FLIP to T cell activation. J Biol Chem 2004; 279: 13–18.
Chuang YT, Lin YC, Lin KH, Chou TF, Kuo WC, Yang KT et al. Tumor suppressor death-associated protein kinase is required for full IL-1β production. Blood 2011; 117: 960–970.
Acknowledgements
This work was supported by grant NHRI-EX101-10012SI from the National Health Research Institute, grant NSC101-2321-B001-002 from National Science Council, and an Academia Sinica Investigator Award from Academia Sinica, and Taiwan, R.O.C. We thank Drs. Luk Van Parijs, Gina Costa, and Garry Nolan for critical reagents, Yamin Lin and FACS Core, Sue-Ping Lee and the Confocal Core of the Institute of Molecular Biology, Academia Sinica for cell sorting and confocal microscopy, and Dr. AndreAna Peña for editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by S Nagata
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Wu, YH., Kuo, WC., Wu, YJ. et al. Participation of c-FLIP in NLRP3 and AIM2 inflammasome activation. Cell Death Differ 21, 451–461 (2014). https://doi.org/10.1038/cdd.2013.165
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2013.165
Keywords
This article is cited by
-
Absent in melanoma 2 (AIM2) in rheumatoid arthritis: novel molecular insights and implications
Cellular & Molecular Biology Letters (2022)
-
How location and cellular signaling combine to activate the NLRP3 inflammasome
Cellular & Molecular Immunology (2022)
-
NLRP3 inflammasome activation and cell death
Cellular & Molecular Immunology (2021)
-
Src-family kinase-Cbl axis negatively regulates NLRP3 inflammasome activation
Cell Death & Disease (2018)
-
Inflammasomes, the eye and anti-inflammasome therapy
Eye (2018)