Abstract
Wnt reception at the membrane is complex and not fully understood. CD44 is a major Wnt target gene in the intestine and is essential for Wnt-induced tumor progression in colorectal cancer. Here we show that CD44 acts as a positive regulator of the Wnt receptor complex. Downregulation of CD44 expression decreases, whereas CD44 overexpression increases Wnt activity in a concentration-dependent manner. Epistasis experiments place CD44 function at the level of the Wnt receptor LRP6. Mechanistically, CD44 physically associates with LRP6 upon Wnt treatment and modulates LRP6 membrane localization. Moreover, CD44 regulates Wnt signaling in the developing brain of Xenopus laevis embryos as shown by a decreased expression of Wnt targets tcf-4 and en-2 in CD44 morphants.
Similar content being viewed by others
Main
Wnt/β-catenin signaling is a key morphogenic pathway regulating numerous developmental processes, including body axis formation, somitogenesis, limb and brain development in multiple organisms (reviewed in Logan and Nusse1). Wnt signaling is crucial during embryogenesis and remains essential throughout life given its essential role in stem cell self-renewal in adult stem and progenitor cells (reviewed in Clevers2). Wnts are lipid-modified glycoproteins binding both Frizzled (Fz) and low-density lipoprotein receptor-related proteins (LRP5/6) (reviewed in Clevers and Nusse3). A hallmark of the activated Wnt signaling is the accumulation of the transcriptional co-activator β-catenin in the nucleus. In the absence of Wnts, β-catenin is phosphorylated, ubiquitinated and targeted for degradation by a cytoplasmic destruction complex containing APC (adenomatous polyposis coli), Axin and the kinases glycogen synthase kinase 3β and casein kinase 1α. In the presence of Wnts, inhibition of the destruction complex results in stabilization of β-catenin, which then translocates to the nucleus, binds to T-cell factor (TCF)/lymphoid enhancer factor (LEF) transcription factors and activates gene transcription (reviewed in Logan and Nusse1).
Prominent Wnt/β-catenin target genes include the proto-oncogene C-MYC,4 the negative Wnt regulator AXIN25,6 and cluster of differentiation 44 (CD44).7 CD44 transmembrane glycoproteins control growth, survival, differentiation and motility (reviewed in Naor et al.8 and Ponta et al.9). The heterogeneity of the CD44 family is generated by alternative splicing of 10 variant exons (v1–v10) in the extracellular domain. These can either be excised completely as in CD44s or be included in various combinations to encode CD44 variant proteins (CD44v).
CD44 controls signaling through interactions with other cell surface receptors such as receptor tyrosine kinases (RTKs), including Met and vascular endothelial growth factor receptor-2 (reviewed in Orian-Rousseau10) and G-protein coupled receptors, such as C-X-C chemokine receptor type 4.11
The key function of CD44 in growth control and in transformation prompted us to test its potential role as a regulator of the Wnt pathway. CD44 is a major Wnt target in the intestine. Accordingly, Tcf4−/− mice show a complete absence of CD44 in the intestinal epithelium, indicating that β-catenin/TCF signaling controls CD44 expression.7 Consistently, the adenoviral expression of Dickkopf1 (Dkk1), a negative regulator of Wnt signaling, represses intestinal CD44 expression in adult mice.12 CD44v6 expression is restricted to proliferating tissues, including the intestinal crypts where the Wnt cascade controls cell fate.7 Recently, CD44v4-v10 isoforms were identified in Leucine-rich repeat-containing G-protein coupled receptor 5 positive (Lgr5+) intestinal stem cells, whereas CD44s was present in transit amplifying cells,13 suggesting a tight regulation of alternative splicing in the intestine. The expression pattern of some CD44 isoforms is shown in Figure 1.7,10,13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23
Expression pattern of CD44 isoforms. All CD44 isoforms are encoded by one single gene present on chromosome 11 in humans and on chromosome 2 in mice. The CD44 gene is composed of 20 exons designated as constant and variable exons. Exons 1–5 and 16–20 are constant exons that encode the N-terminal and C-terminal regions of CD44, respectively. Exon 19 is, however, spliced out in most CD44 isoforms. Ten variant exons (v1–v10) encoded by exons 6–15 can be alternatively spliced and included in various combinations in the stem region, thereby giving rise to many CD44 variant isoforms. In human cells, exon v1 (exon 6) encodes a stop codon and is not expressed. The smallest CD44 isoform, CD44s, does not contain any variant exon in the stem region and is virtually ubiquitously expressed. In contrast, the expression of CD44 variant isoforms is restricted to specific tissues
Overexpression of CD44v6 correlates with advanced stages of colorectal cancer24 characterized by mutations in the Wnt pathway (e.g., APC mutations). In the ApcMin/+ mouse (mutation at codon 850), CD44 controls the onset of tumorigenesis as deduced by a 50% reduction of intestinal adenomas in ApcMin/+/Cd44−/− mice.25 Moreover, inhibition of CD44v6 by means of shRNA reduced the number of adenomas in ApcMin/+ mice.26
Besides the well-described function of CD44 as a Wnt/β-catenin target gene, little is known about the role of CD44 as a potential modulator of the pathway itself. CD44 downregulation was recently shown to affect β-catenin phosphorylation and nuclear accumulation in chronic myeloid leukemia cells.27 Induction of the canonical Wnt pathway with specific activators such as Wnt ligands was, however, not tested. As β-catenin can be activated by several pathways,28, 29, 30 the requirement of CD44 for the Wnt-dependent β-catenin activation remains to be addressed.
Here we show that CD44 acts as a regulator of the Wnt-induced β-catenin signaling pathway. Downregulation of CD44 decreases, whereas overexpression of CD44 isoforms, increases Wnt/β-catenin signaling. Epistasis experiments place CD44 function at the receptor level, regulating LRP6 phosphorylation and localization at the membrane. To influence the Wnt pathway, CD44 requires the binding of its intracellular cytoplasmic domain (ICD) to ERM (Ezrin-Radixin-Moesin) and to the cytoskeleton. Most importantly, the expression of the Wnt target genes tcf-4 and engrailed-2 (en-2) in Xenopus laevis is decreased upon injection of CD44 morpholinos (MOs), indicating a role of CD44 in Wnt-dependent patterning of the central nervous system (CNS).31,32
Results
CD44 is a positive regulator of canonical Wnt signaling
We first investigated whether repression of CD44 expression by RNAi would alter Wnt3a-regulated gene expression. For Wnt pathway activation, we used the Wnt3a conditioned medium (Wnt3a-CM, designated Wnt3a). We transfected HEK293 cells expressing several CD44 isoforms33 with the TCF/LEF responsive reporter gene construct TOPFlash and siRNA targeting all CD44 isoforms. Treatment with Wnt3a resulted in activation of Wnt signaling, and this was significantly reduced by CD44 siRNA (Figure 2a). Conversely, overexpression of human CD44s led to a dose-dependent increase of Wnt signaling (Figure 2b). These results suggest that CD44 can positively regulate Wnt/β-catenin signaling.
CD44 is a positive regulator of Wnt/β-catenin signaling. (a) HEK293 cells transfected with control siRNA or siRNA against all CD44 isoforms were transfected with TOPFlash and control TK-Renilla vectors. After stimulation with Wnt3a-CM or Co-CM (20 h), cells were lysed and subjected to luciferase measurements or, in parallel, to western blotting (WB) analysis. Insert: siRNA-transfected HEK293 cells were transfected with FOPFlash and control TK-Renilla vectors and 48 h later subjected to luciferase measurements. (b) HEK293 cells were transfected with TOPFlash and control TK-Renilla vectors together with increasing amounts of hCD44s cDNAs. After stimulation with Wnt3a-CM or Co-CM (20 h), cells were lysed and subjected to luciferase measurements or, in parallel, to WB analysis. Insert: HEK293 cells were transfected with an empty vector or hCD44s (75 ng) together with FOPFlash and control TK-Renilla vectors and 48 h later subjected to luciferase measurements. (c) HeLa cells were transfected with control siRNA or siRNA against all CD44 isoforms and treated either with Co-CM or Wnt3a-CM for 3 h. Cells were subjected to WB analysis 72 h after siRNA transfection. The numbers indicate the fold intensity values of the ABC bands normalized to the actin bands. (d) HeLa cells were transfected with control-siRNA or siRNA against all CD44 isoforms. Forty-eight hours after siRNA transfection, cells were treated with Co-CM or Wnt3a-CM for 24 h. Subsequently, cells were subjected to immunofluorescence staining using antibodies against β-catenin (red). The nucleus was stained with DAPI (4,6-diamidino-2-phenylindole; blue). Afterwards, cells were analyzed by confocal microscopy (left panel: β-catenin only; right panel: overlay; scale bar=10 μm), and the percentage of cells with nuclear β-catenin was evaluated (diagram). Data represent mean±S.D. of all confocal microscopy experiments (four independent experiments; n>25 cells per experiment were evaluated). Statistical significance was analyzed using the Student’s t-test (*P<0.05). The knockdown of CD44 was confirmed by WB analysis. (e) HEK293 cells were transfected with TOPFlash and control TK-Renilla vectors together with 75 ng of rCD44v4-v7, rCD44v6 or rCD44s cDNAs. After stimulation with Wnt3a-CM or Co-CM (20 h), cells were lysed and subjected to luciferase measurements or, in parallel, to WB analysis. Reporter gene assays and WB analysis are described in ‘Materials and Methods’. All TOPFlash data represent mean±S.D. from at least four independent experiments performed in triplicates. Statistical significance was analyzed using the Student’s t-test (*P<0.05, ***P<0.005)
We next measured the levels of hypophosphorylated, active β-catenin (ABC), as a read-out for pathway activity in HeLa cells that express various CD44 isoforms.34 Wnt3a-induced β-catenin activation in these cells was clearly inhibited by CD44 siRNA (Figure 2c), confirming the requirement of CD44 for Wnt3a-induced Wnt/β-catenin signaling. A similar effect was also seen in HCT116 cells (Supplementary Figure S1A). In agreement, cell-fractionation experiments in HeLa cells showed a reduction of the Wnt-induced cytosolic accumulation of β-catenin by CD44 siRNA (Supplementary Figure S1B). Additional evidence was obtained using β-catenin translocation to the nucleus in HeLa cells as a read out. Indeed, immunofluorescence analysis demonstrated that nuclear translocation was drastically blocked in cells transfected with CD44 siRNA (Figure 2d).
CD44v isoform expression correlates with areas of high Wnt activity. In colorectal cancer, for example, expression of v6-containing isoforms strongly correlates with tumor progression,24 which is largely controlled by the Wnt pathway (reviewed in Radtke and Clevers35). In addition, the intestinal Lgr5+ stem cells are CD44v4-v10 positive, suggesting a role of variant isoforms of CD44 in intestinal stem cell fate.13 To investigate whether CD44v isoforms have specific effects on Wnt signaling, we compared the rat variant isoforms CD44v4-v7, CD44v6 and CD44s in the TOPFlash assay (Figure 2e). All CD44 isoforms augmented Wnt3a-activated transcription to the same extent, suggesting that the inclusion of variant exons is not necessary for CD44’s influence on canonical Wnt signaling. These results suggest that all CD44 isoforms function as positive regulators of Wnt signaling.
All CD44 proteins bind hyaluronan (HA) through their extracellular domains and ERMs through their ICDs (reviewed in Ponta et al.9). CD44-mediated amplification of Wnt/β-catenin signaling is, however, independent of HA. Indeed, hyaluronidase treatment or antibodies blocking the HA/CD44 binding had no effect on Wnt signaling (Supplementary Figures S2A and B). Concordantly, overexpression of a CD44 mutant lacking the HA binding ability36 was as efficient as overexpression of wild-type CD44 in activating Wnt signaling (Supplementary Figure S2C).
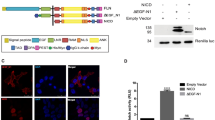
The CD44 ICD binds to several signaling molecules, including ERMs.37 Therefore, we investigated the role of the CD44 ICD in Wnt regulation. We expressed a rat CD44v4-v7 full-length protein or a rat CD44 mutant lacking the ICD (rCD44v4-v7ΔCyt) and tested their ability to amplify Wnt signaling in HEK293 cells (Figure 3a). The rat CD44 proteins can be detected by western blotting using a rat-specific antibody, and these constructs have already been used to study signal transduction.38 In contrast to full-length rat CD44v4-v7, rCD44v4-v7ΔCyt, a membrane-localized protein (Supplementary Figure S3A), did not increase Wnt3a-induced TOPFlash activation, demonstrating that the effect of CD44 on Wnt/β-catenin signaling is mediated through the ICD.
CD44 requires the ICD and its binding to F-actin via Ezrin to regulate Wnt signaling. (a) HEK293 cells were transfected with TOPFlash and control TK-Renilla vectors together with an empty vector or cDNAs for rCD44v4-v7 or rCD44v4-v7ΔCyt (75 ng). After stimulation with Wnt3a-CM or Co-CM (20 h), cells were lysed and subjected to luciferase measurements or, in parallel, to WB analysis. (b) HEK293 cells were transfected with TOPFlash and control TK-Renilla vectors together with empty vector or cDNAs for GST-tagged CD44-Cyt (CD44 ICD, 75 ng) or GST-tagged CD44-CytΔEzBD (CD44 ICD mutated in the Ezrin binding domain, 75 ng). After stimulation with Wnt3a-CM or Co-CM (20 h), cells were lysed and subjected to luciferase measurements or, in parallel, to WB analysis. (c) HeLa cells were transfected either with empty vector or cDNAs for CD44-Cyt or CD44-CytΔEzBD, treated with Wnt3a-CM for 3 h and subjected to WB analysis. (d) HEK293 cells transfected with control siRNA or siRNA against Ezrin were transfected with TOPFlash and control TK-Renilla vectors. After stimulation with Wnt3a-CM or Co-CM (20 h), cells were lysed and subjected to luciferase measurements or, in parallel, to WB analysis. (e) HEK293 cells were transfected with TOPFlash reporter and control Renilla luciferase vectors together with an empty vector or a cDNA corresponding to a DN V-SVG-tagged mutant of Ezrin (DN-Ezrin, 20 ng) mutated in the F-actin binding domain. After stimulation with Wnt3a-CM or Co-CM (20 h), cells were lysed and subjected to luciferase measurements or, in parallel, to WB analysis. Reporter gene assays and WB analysis are described in ‘Materials and Methods’. All data represent mean±S.D. from at least four independent experiments performed in triplicates. Statistical significance was analyzed using the Student’s t-test (*P<0.05; n.s.=not significant)
CD44 requires the binding to Ezrin (Ez) to its ICD for its function in Wnt/β-catenin signaling
Previous work showed a dominant-negative (DN) effect of the CD44 ICD,38 so we tested whether its overexpression would block Wnt signaling. Indeed, overexpression of the CD44 ICD (CD44-Cyt) significantly inhibited Wnt3a-induced TOPFlash activity (Figure 3b) as well as β-catenin activation (Figure 3c). Taken together, these data show that the ICD of CD44 mediates its function in Wnt signaling.
We addressed the role of ERMs in CD44-dependent regulation of Wnt signaling using a CD44 ICD construct mutated in the ERM-binding site (CD44-CytΔEz-BD). Unlike the wild-type ICD, the CD44-CytΔEz-BD protein showed no significant effect on Wnt3a-induced TOPFlash activation (Figure 3b) or β-catenin activation (Figure 3c). Therefore, the CD44 ICD requires association with ERM proteins for its function in Wnt signaling. Concordantly, a CD44 full-length molecule mutated in the ERM binding domain was not as efficient as wild-type CD44 in increasing Wnt signaling in a TOPFlash assay (Supplementary Figure S3B).
The role of ERMs in Wnt signaling was directly addressed in loss-of-function experiments in HEK293 cells using both siRNA-mediated knockdown of Ezrin and overexpression of an Ezrin DN mutant lacking the actin-binding site (Figures 3d and e).39 Similar to RNAi-mediated CD44 knockdown, both Ezrin RNAi and DN-Ezrin strongly suppressed Wnt3a-mediated activation of the Wnt reporter. This confirms the involvement of Ezrin in Wnt signaling and suggests that its link to the cytoskeleton is required for CD44-mediated Wnt signaling.
CD44v6 regulates RTK signaling (reviewed in Orian-Rousseau10). RTKs induce LRP6 phosphorylation and β-catenin signaling via the mitogen-activated protein kinase (MAPK)/Erk and phosphatidylinositol 3 kinase (PI3K)/Akt pathways.29 However, inhibitors of both pathways did not influence CD44-dependent augmentation of Wnt signaling (Supplementary Figures S4A and B).
CD44 acts at the level of the Wnt receptors
Epistasis experiments in the context of CD44 loss- and gain-of-function were performed to identify CD44-specific targets in the Wnt signaling cascade (Figure 4). In the loss-of-function experiment, cells were treated either with control or CD44 siRNA. Wnt/β-catenin signaling was activated using either Wnt3a or overexpression of LRP6, dishevelled (Dsh) or constitutive ABC (Figure 4a). In addition, gain-of-function epistasis experiments using overexpression of CD44s rather than siRNA treatment (Figure 4b) were performed. Strikingly, both approaches showed a clear involvement of CD44 in Wnt signaling when the pathway was activated by Wnt3a or LRP6 but not by the cytoplasmic Wnt transducers Dsh or β-catenin (Figures 4a and b). The most dramatic effect of CD44 manipulation was seen on LRP6-induced Wnt signaling, suggesting that CD44 regulates Wnt signaling at the level of membrane receptors.
CD44 regulates Wnt signaling at the level of the membrane. (a) HEK293 cells were transfected with control siRNA or siRNA against all CD44 isoforms. Twenty hours later, cells were transfected with TOPFlash and control TK-Renilla vectors. The reporter was activated either by treatment with Wnt3a-CM or by co-transfection of cDNAs for LRP6 (20 ng), Dsh (20 ng) or constitutive ABC (3 ng). Forty hours after DNA transfection, cells were lysed and subjected to luciferase measurements or, in parallel, to WB analysis. (b) HEK293 cells were transfected with TOPFlash and control TK-Renilla vectors together with empty vector or a cDNA for rCD44s (75 ng). The reporter was activated either by treatment with Wnt3a-CM or by co-transfection of LRP6 (20 ng), Dsh-1 (20 ng) or constitutive ABC (3 ng). Forty hours after transfection, cells were analyzed for luciferase activity. Forty hours after transfection, cells were lysed and subjected to luciferase measurements or, in parallel, to WB analysis. Reporter gene assays and WB analysis are described in ‘Materials and Methods’. All data represent mean±S.D. from at least four independent experiments performed in triplicates. Statistical significance was analyzed using the Student’s t-test (*P<0.05; n.s.=not significant)
CD44 associates with LRP6 and is required for LRP6 activation and membrane localization
A possible association between CD44 and LRP6 was investigated in co-immunoprecipitation (co-IP) studies (Figures 5a and b) using HeLa cells. We first overexpressed CD44v4-v7 and FLAG-LRP6 in control or Wnt3a-treated cells. Lysates were then immunoprecipitated with a CD44 antibody, and potential co-immunoprecipitation of LRP6 was tested using an anti-FLAG antibody. Strikingly, interaction between CD44v4-v7 and LRP6 was only detected in the presence of Wnt3a (Figure 5a). A CD44v4-v7 mutant lacking the ICD (CD44ΔCyt) did not associate with LRP6, indicating that the CD44 ICD is required for its Wnt-induced association with LRP6 (Figure 5a). This finding might explain why the CD44ΔCyt, in contrast to the full-length CD44, did not increase Wnt3a-induced TCF/LEF-regulated transcription (Figure 3a). We next tested for Wnt-induced association of endogenous CD44 and LRP6. HeLa cells were incubated with Wnt3a for different time points, lysates treated with a CD44 antibody and immunoprecipitates tested for the presence of LRP6. As expected, no LRP6 was seen in untreated cells; however, a clear band corresponding to LRP6 was apparent in CD44 immunoprecipitates already after 5 min of Wnt3a treatment (Figure 5b). Thus CD44 and LRP6 interact in a Wnt-dependent manner. The co-immunoprecipitation between FLAG-LRP6 and endogenous CD44 was confirmed in the other direction (Supplementary Figure S5A). The association between CD44 and LRP6 depends most likely on the CD44-cytoskeleton linkage through ERMs. Indeed, in contrast to a full-length wild-type CD44, a mutant of CD44 in the ERM-binding domain was not found in complex with LRP6 (Supplementary Figure S5B). In addition, transfection of a DN form of Ezrin, lacking the actin-binding domain, prevented the Wnt-dependent complex formation between CD44 and LRP6 (Figure 5c). The CD44–LRP6 interaction occurs through the respective cytoplasmic domains. Indeed, a glutathione S-transferase (GST)–CD44 cytoplasmic domain interacted with a FLAG-LRP6 mutant lacking the extracellular domain (LRP6ΔE(1–4)40) in HeLa cells (Supplementary Figure S5C).
CD44 interacts with LRP6 and is required for LRP6 activation. (a) HeLa cells were transfected with cDNAs for rCD44v4-v7 or rCD44v4-v7ΔCyt together with a FLAG-tagged LRP6 construct. Twenty-four hours after transfection, cells were treated either with Co-CM or Wnt3a-CM for 30 min, lysed and proteins were immunoprecipitated with an antibody against rCD44. Immunoprecipitates were subjected to WB analysis using the FLAG and rCD44 antibodies. (b) HeLa cells were treated for the indicated time points with Wnt3a-CM, lysed and proteins were immunoprecipitated with antibodies against CD44. Immunoprecipitates were subjected to WB analysis with an antibody against LRP6 and CD44. The numbers indicate the fold increase of co-immunoprecipitated LRP6 normalized to the immunoprecipitated CD44. (c) HeLa cells were transfected with cDNAs for DN-Ezrin together with a FLAG-tagged LRP6 construct. Twenty-four hours after transfection, cells were treated either with Co-CM or Wnt3a-CM for 30 min, lysed and proteins were immunoprecipitated with an antibody against hCD44. Immunoprecipitates were subjected to WB analysis using the FLAG and hCD44 antibodies. (d) HeLa cells were transfected with control siRNA or siRNA against all CD44 isoforms and treated either with Co-CM or Wnt3a-CM for 2 h. Cells were subjected to WB analysis 72 h after siRNA transfection (asterisks=mature LRP6 band of untreated (red) or Wnt3a treated (green) cells). The numbers indicate the fold intensity values of the p-LRP6 bands normalized to the bands of total LRP6
We specifically tested the involvement of CD44 in LRP6 phosphorylation at Serine 1490 (Ser1490), an early event in the activation of the Wnt signaling cascade41 (Figure 5d). Indeed, siRNA-mediated knockdown of CD44 in HeLa cells strongly inhibited Wnt3a-induced phosphorylation of LRP6 as measured with a phospho-Ser1490 antibody (Figure 5d).
LRP6 is usually detected as two bands with different molecular weights in SDS-PAGE/WB. The immature form of LRP6 (faster migrating band) is located in the endoplasmic reticulum (ER), whereas the mature (upper band) more heavily glycosylated form is at the plasma membrane.42 Upon Wnt activation, the upper band is further up-shifted due to a higher phosphorylation status. Consistent with the loss of PPSP phosphorylation, this Wnt3a-induced up-shift of LRP6 is reduced upon CD44 knockdown (green asterisks in Figure 5d). Moreover, loss of CD44 also lead to reduction of the upper band of endogenous LRP6 in unstimulated cells (red asterisks in Figure 5d) suggesting that CD44 drives the maturation of LRP6 or its localization at the membrane (Figure 5d, total LRP6 panel).
The localization of LRP6 at the membrane, where LRP6 phosphorylation takes place, was tested upon overexpression of green fluorescent protein (GFP)-tagged LRP6 in HeLa cells transfected with either control or CD44 siRNA (Figure 6a). Whereas 80% of control siRNA-transfected cells displayed LRP6-GFP at the membrane (Figure 6a), in 75% of CD44 siRNA-transfected cells, most LRP6 was perinuclear (Figure 6a). Correct membrane localization of LRP6 could be restored in the CD44 siRNA-transfected cells upon transfection of rat CD44s (Figure 6a). Here LRP6 was found again at the membrane in 74% of cells. This effect was specific for LRP6, because lack of CD44 did not affect receptor tyrosine kinase-like orphan receptor 2 (Ror2) membrane localization (Figure 6a). Further analysis of LRP6 localization in the absence of CD44 revealed that the majority of LRP6 is retained in the ER as demonstrated by the colocalization of LRP6 with a specific ER marker (Figure 6b). The staining of LRP6-GFP at the plasma membrane was slightly enhanced upon overexpression of the LRP5/6-specific chaperone mesoderm development (Mesd) that promotes transit of LRP6 from the ER to the membrane42 (Figure 6b). However, even in the presence of co-expressed Mesd, LRP6 although released from the ER still failed to reach the plasma membrane in CD44 siRNA-transfected cells (Figure 6b). LRP6 membrane localization was also strongly suppressed by transfection of the cytosolic CD44 ICD (Figure 6c) acting in a DN manner. Indeed, in only 30% of ICD transfected cells LRP6 was membrane localized. The inhibition of LRP6 membrane localization by the CD44 ICD was rescued by mutation of the Ezrin-binding sites, suggesting a role of Ezrin in this process. Moreover, the Ezrin–cytoskeleton linkage is required, as a DN-Ezrin also blocked the correct LRP6 localization (Figure 6c). Therefore, the function of CD44 in Wnt signaling strongly depends on its ICD and its binding to the cytoskeleton via Ezrin.
Membrane targeting of LRP6 is dependent on CD44 and its binding to F-actin via Ezrin. (a) HeLa cells transfected either with control siRNA or siRNA against all CD44 isoforms were transfected with LRP6-GFP or Ror2-mCherry constructs, and the cellular localization of LRP6-GFP/Ror-mCherry was determined using a Leica SP5 fluorescence microscope with a 63X objective (scale bar=10 μm). Where indicated, in addition to hCD44 siRNA, rCD44s was co-transfected. Quantification is shown in the histogram. (b) HeLa cells transfected either with control siRNA or siRNA against all CD44 isoforms were transfected with a construct for LRP6-GFP and counterstained with an ER marker. Where indicated, cells were co-transfected with Mesd cDNA. Localization of LRP6-GFP was detected as in panel (a). In addition, the percentage of cells with colocalized LRP6-GFP and ER marker was evaluated. (c) HeLa cells transfected either with an empty vector or constructs for CD44 ICD (CD44-Cyt), the CD44 ICD mutated in the Ezrin binding domain (CD44-CytΔEzBD) or DN-Ezrin together with a construct for LRP6-GFP. LRP6-GFP was detected as in panel (a). Data represent mean±S.D. of all confocal microscopy experiments (at least four independent experiments, n>30 cells per condition were evaluated, scale bar=10 μm). Statistical significance was analyzed using the Student’s t-test (*P<0.05; n.s.=not significant)
CD44 is required for Wnt target gene expression in the developing X. laevis CNS
The physiological relevance of CD44 function in Wnt signaling was tested in X. laevis. Overexpression of CD44s from human, rat or X. laevis in HEK293 cells enhanced Wnt3a-induced signaling to a similar extent (Figure 7a), demonstrating a conserved function of CD44 as a Wnt regulator across species.
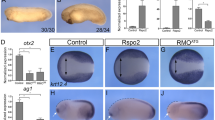
CD44 is required for Wnt target gene expression in Xenopus laevis. (a) HEK293 cells were transfected with TOPFlash and control TK-Renilla vectors together with 75 ng of cDNAs for hCD44s, rCD44s or xCD44s. After stimulation with Wnt3a-CM or Co-CM (20 h), cells were lysed and subjected to luciferase measurements. Data represent mean±S.D. from at least four independent experiments performed in triplicates. Statistical significance was analyzed using the Student’s t-test (*P< 0.05). (b) In situ hybridization for the canonical Wnt target genes tcf-4 and en-2 in 32-stage X. laevis embryos. Embryos were injected in the right blastomere at two-cell stage with Co-MO, xCD44-MO alone, xCD44-MO together with cDNA for hCD44s expression vectors or xCD44-MO together with β-catenin expression vectors. FITC-labeled Dextran was co-injected to assess the side of injection. (c) Quantification of experiments described in panel (b). Data represent mean±S.D. from at least four independent experiments; n>75 per condition were evaluated; statistical significance was analyzed using the Student’s t-test (*P<0.05)
Expression of the Wnt target genes tcf-4 and en-2, both expressed in the developing brain, along with CD4443 was measured upon downregulation of CD44 using MO antisense oligonucleotides. Examination of stage-32 embryos showed that the CD44-MO worked (Supplementary Figure S6). Indeed, MO-injected embryos failed to develop a ventral musculature as already shown in Ori et al.,31,43 a phenotype rescued by injection of human CD44s DNA (Supplementary Figure S6). We injected CD44-MO in one blastomere of X. laevis two-cell stage embryos and tested the consequence on the expression levels of tcf-4 and en-2 transcripts using the whole-mount in situ hybridization. Expression of both target genes was significantly reduced on the injected side in >30% of injected embryos (Figures 7b and c). Importantly, co-injection of hCD44s DNA partially rescued this phenotype (Figures 7b and c), confirming that the reduced expression of the Wnt target genes is specific for the CD44 knockdown. Injection of β-catenin DNA also rescued this phenotype, showing that downregulated expression of tcf-4 and en-2 by CD44-MO is indeed specific to Wnt/β-catenin signaling (Figures 7b and c). These results provide compelling evidence for the in vivo relevance of CD44 function in Wnt signaling. Moreover, as tcf-4 is a marker for the midbrain44 and en-2 defines the midbrain/hindbrain boundary,45 this result indicates an involvement of CD44 in CNS patterning during early embryogenesis.
Taken together, our data suggest a novel and essential role for CD44 in Wnt signaling by regulating the cell surface expression and signaling activity of the Wnt/β-catenin co-receptor, LRP6.
Discussion
We describe here a novel function of CD44 in Wnt signaling. Our findings indicate that CD44 is not only a Wnt target gene but also functions as a modulator of Wnt/β-catenin signaling at the level of LRP6 activation and membrane localization. Whereas downregulation of CD44 leads to decreased β-catenin activation and TCF/LEF reporter gene activation, overexpression of CD44 increases Wnt activity (Figure 2), indicating a function as a positive regulator of Wnt signaling. CD44 associates with LRP6 upon Wnt stimulation (Figure 5) and is also required for correct LRP6 localization at the membrane (Figure 6). Our findings indicate that CD44 is a positive feedback regulator of the Wnt pathway, in line with the idea that feedback control is a key feature of Wnt signaling regulation (reviewed in Logan and Nusse1). The in vivo relevance of these findings is demonstrated in the developing X. laevis embryo where the Wnt-dependent anterior–posterior patterning of the X. laevis brain requires CD44 (Figure 7).
Epistasis experiments (Figure 4) demonstrate that CD44 acts at the level of LRP6 and upstream of Dsh and β-catenin. The fact that CD44 is also required for Wnt-dependent LRP6 phosphorylation further supports this (Figure 5d). These data are in agreement with the idea that LRP6 lies genetically upstream of Dsh.46 In contrast to this assumption, however, Dsh was shown to be required for LRP6 phosphorylation, a function that would place it biochemically upstream of LRP6 activation.47 Interestingly, CD44 has a role not only in Wnt-dependent phosphorylation of LRP6 but also in LRP6 translocation to the membrane. Therefore, a possible explanation for the lack of Wnt-dependent phosphorylation of LRP6 (Figure 5d) in the absence of CD44 might be the inhibition of LRP6 translocation to the membrane. However, LRP6 activation is a Wnt-dependent process, whereas LRP6 transport is Wnt independent. Furthermore, CD44 interacts with LRP6 only upon Wnt stimulation, whereas the function of CD44 in LRP6 transport to the membrane seems to be more indirect. Therefore we suggest that these are two independent functions of CD44 in Wnt signaling.
The ERM-binding motifs are required for CD44-dependent Wnt regulation (Figure 3b,Supplementary Figure S3B), suggesting that the binding of the CD44 ICD to ERMs is involved. Furthermore, we show here that Ezrin binding to the cytoskeleton has a decisive role in this pathway (Figure 3e). Ezrin function might be linked to the formation of LRP6 signalosomes via cytoskeleton rearrangements, a step crucial for Wnt signaling. Indeed, actin–cytoskeletal rearrangements have already been shown to control Wnt/β-catenin signaling in mesenchymal cells.48 Moreover, the CD44 ICD and the Ezrin actin-binding domain control LRP6 membrane localization, suggesting that the transport of vesicles to the membrane involves actin polymerization. This latter step is promoted by the actin-related proteins 2/3 (Arp2/3) complex and neural Wiskott–Aldrich syndrome protein (N-WASP). It was shown that CD44 and Ezrin interact with N-WASP and promote the actin assembly via the N-WASP-Arp2/3 machinery.30,49
Altogether, the role of CD44 both in the Wnt-independent LRP6 membrane targeting and in the Wnt-dependent LRP6 phosphorylation appear to be linked to the ability of CD44 to bind ERMs. Functions of ERMs that might explain their role in these two processes have been described. On the one hand, ERMs regulate membrane delivery of proteins.50 On the other hand, ERMs function as scaffold proteins that build molecular networks facilitating activation of proteins.51
Our results show that CD44 is a positive regulator of Wnt signaling. This is in line with the fact that the Wnt pathway is controlled by feedback mechanisms. Negative feedback loop mechanisms are understandably necessary to avoid uncontrolled activation of the Wnt pathway. In contrast, positive regulatory mechanisms are less common. One possible target organ for positive regulation is the intestine. Indeed, the consequences of an abrogation of canonical Wnt signaling by either removal of β-catenin or TCF4 or by overexpression of the Wnt inhibitor Dkk1 are dramatic. Inactivation of canonical Wnt signaling caused in all cases complete loss of proliferation of intestinal progenitor cells at the bottom of the intestinal crypts and had severe effects on stem cell maintenance and tissue homeostasis.12,52,53 In this tissue in particular, an isoform-specific expression of CD44 was shown. Indeed, whereas CD44v4-v10 is expressed in intestinal stem cells, proliferative progenitor cells express CD44s.13 A positive feedback of CD44 might be crucial for the maintenance of a constant level of Wnt activity required for the homeostasis and renewal of the intestinal epithelium.
It has been extensively shown that CD44 is involved in Wnt-induced tumorigenesis.7 Till now, only the aspect of CD44 as Wnt target gene has been investigated. The finding that CD44 is not only a Wnt target gene but also a Wnt pathway regulator sheds new light on the role of CD44 in Wnt-induced tumorigenesis.
Materials and Methods
Cell culture
All cell lines (HEK293 CRL-1573; HeLa CCL-2; Wnt3a-L cells CRL-2647; L-cells CRL-2648; HCT116 CCL-247) were obtained from ATCC (Wesel, Germany) and were maintained in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum (FBS). Wnt induction was done using recombinant Wnt3a (200 ng/ml; R&D Sytems, Wiesbaden, Germany) and mouse Wnt3a-CM, produced from mouse L cells stably transfected with mouse Wnt3a.54 As Control conditioned medium (Co-CM), we used medium from L-cells. The Wnt3a-CM was diluted in DMEM+10% FBS to a concentration that induced the TOPFlash reporter comparably to 200 ng/ml recombinant Wnt3a in DMEM+FBS. U0126 and LY294002 were purchased from Cell Signaling (Beverly, UK).
Antibodies and other reagents
The human pan-CD44 antibody Hermes 3 was a gift from S Jalkannen (Turku, Finland); the ratCD44 exon 15-specific antibody 5G8 and the rat exon v6-specific antibody 1.1ASML have been described.55,56 The human CD44 antibody BU52 was obtained from Pierce (Rockford, IL, USA) and is described previously.57 The mouse IgG1 antibodies came from R&D Systems. The anti-ABC antibody clone 8E7 was obtained from Millipore (Bedford, MA, USA), and the antibody against total β-catenin used for western blotting analysis was from BD Transduction Laboratories (Mat. no.: 610153; Heidelberg, Germany). The antibody against total β-catenin (D10A8) used for immunofluorescence microscopy and the Phospho-LRP6 (Ser1490) antibody were obtained from Cell Signaling. The total LRP6 antibody T1479 was described previously.40 The anti-V-SVG antibody used for detection of V-SVG tagged DN Ezrin was from Stressgen (Ann arbor, MI, USA). Antibodies against Ezrin (3C12), actin (I-19), PCNA (PC10), ERK1/2 (K-23) and GST (Z-5) were purchased from Santa Cruz (Heidelberg, Germany). The antibody against the transferrin receptor (Ab84036) was obtained from Abcam (Cambridge, UK). The anti-FLAG antibody (Clone M2), anti-FLAG beads (Clone M2) and GSH gluthatione agarose (G4510) were from Sigma (Taufkirchen, Germany). Mouse IgG-sepharose bead conjugates (Cell Signaling) were used as controls for the co-immunoprecipitation experiments. Anti-mouse or anti-rabbit horseradish peroxidase-coupled secondary antibodies were from Dako (Hamburg, Germany). Hyaluronan (Healon) was from Abbott Medical Optics (Santa Ana, CA, USA). The MAPK kinase (MEK) inhibitor UO126 was purchased from Promega (Mannheim, Germany). The PI3K inhibitor Ly249002 was from Cell Signaling. The DMSO used as control for the MEK- and PI3K-inhibition experiments was obtained from Fluka (Neu-Ulm, Germany).
Plasmids and constructs
The human and rat CD44s were cloned in pcDNA3.1 (+/−) (Invitrogen, Karlsruhe, Germany). Sequences are available upon request. xCD44s (X. laevis hypothetical protein LOC443701) cloned into pCMV-Sport6 was a gift from M Ori (University of Pisa, Pisa, Italy). The pGK-CD44v6 and pGK vector controls have been described.58 The sequence of rCD44v4-v7 or rCD44v4-v7ΔCyt cloned into pSV and the pSV empty vector control are described.38 The constructs encoding the CD44 ICD (CD44-Cyt) and the mutant ICD (CD44-CytΔEz-BD) and empty vector control pEBG are described.37 hCD44s mutated in the ERM binding site (CD44s-ERMmut: R292-R293-R293→A292-A293-A294) was generated as previously described.37 hCD44s mutated in the HA-binding site was generated by a point mutation (R41A) in the HA-binding domain and is described previously.36 The construct expressing an Ezrin mutant deleted in the last 29 amino acids at the N-terminus, thus lacking the actin-binding domain (Ezrin-DN) and corresponding empty vector pCB6, are described previously.39 The M50 Super 8x TOPFlash vector (plasmid 12456) with a luciferase gene under the control of seven TCF/LEF-binding sites and the corresponding M51 Super 8x FOPFlash vector (plasmid 12457) with mutated TCF/LEF-binding sites were obtained from Addgene (Cambridge, MA, USA). The normalization vector pRL-TK renilla with a HSV-TK promotor driving Renilla luciferase was purchased from Promega. pCS-FLAG-hLRP6 and pCS-hLRP6-GFP constructs are described previously.59 The pCS2-LRP6 construct is described previously.60 The pCS-FLAG-LRP6ΔE(1-4) construct is described previously.40 hDsh-1 was cloned into pCS2+ and is described previously.61 The myc tagged mβ-Cat expression vector point mutated at Serin 33 (S33A) is described previously.62 The xRor2-mCherry construct is described previously.63 The pCMV-mMesd construct is described previously.40 The xCD44-MO (5′-TAACAATCCACAGCATTGAGGCCAT-3′)43 and standard control-MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′) were obtained from Gene Tools (Philomath, OR, USA). Capped mRNAs were transcribed from linearized DNA templates using mMESSAGE mMachine (Ambion, Warrington, UK). Digoxigenin-labeled antisense probes for in situ hybridization were synthesized with the DIG RNA labeling kit (Roche, Mannheim, Germany). Probes for in situ hybridization are described elsewhere: en-2 in Landesman and Sokol(64), and tcf-4 in Konig et al.65
Transfection procedures
siRNA transfection in HEK293 or HeLa cells was performed using the HiPerFect transfection reagent (Qiagen, Hilden, Germany) according to the manufacturer’s protocol using 5 nM of CD44 pan siRNA (pool of three: (5′-CTGAAATTAGGGCCCAATT-3′; 5′-AATGGTGCATTTGGTGAAC-3′; 5′-CAGAAACTCCAGACCAGTT-3′; Qiagen) or 5 nM Ezrin siRNA (pool of three: 5′-GGAACAUCUCUUUCAAUGA-3′, 5′- CCACGUCUGAGAAUCAACA-3′ and 5′-GACUCUGUUUGCUUGUGUU-3′; Santa Cruz) or control siRNA (pool of three: 5′-UAAUGUAUUGGAACGCAUAUU-3′; 5′-AGGUAGUGUAAUCGCCUUGUU-3′ and 5′-UGCGCUAGGCCUCGGUUGCUU-3′; Eurofins MWG, Ebersberg, Germany). Cells were analyzed at least 48 h after siRNA transfection. DNA transfection of HEK293 and HeLa cells was performed using PromoFectin (PromoKine, Heidelberg, Germany) according to the manufacturer’s protocol. Cells were analyzed at least 24 h after DNA transfection.
Western blotting analysis
HEK293 or HeLa cells were lysed in boiling SDS-sample buffer (125 mMTrisHCl pH 6.8; 4% SDS; 20% Glycerol; 0.01% bromphenol blue) containing 100 mM dithiothreitol (DTT) and subjected to the standard SDS-PAGE and western blotting procedures. Where necessary, blots were stripped (62.5 mM Tris, pH 6.8; 2% SDS; 0.8% DTT; 1 h at 55 °C) and incubated with loading control antibodies. Blots were stained using the enhanced chemiluminescence system (ECL; Thermo Fisher Scientific, Schwerte, Germany). All antibodies used are described above. Where indicated, protein bands were quantified using ImageJ (U.S. National Institutes of Health, Bethesda, MD, USA). Western blot pictures show one representative out of at least three independent experiments.
Hyaluronan detection and hyaluronidase treatment
HEK293 cells were treated for 24 h with 50 U/ml Bovine Testis Hyaluronidase (Sigma), fixed with 4% paraformaldehyde (PFA) and incubated with biotinylated HA-binding protein (Merck, Darmstadt, Germany, 2.5 mg/ml) overnight. Cells were stained with Alexa488–streptavidin conjugates for 2 h (Life Technologies, Darmstadt, Germany) and counterstained with DAPI (Life Technologies). The slides were mounted with Fluorescence Mounting Medium (Dako) and analyzed with a SP5 laser scanning confocal microscope (Leica, Wetzlar, Germany) with a × 63 objective.
Reporter assays
All reporter gene assays were performed in 96-well plates. Briefly, HEK293 cells (1.5 × 104/well) were transfected with SuperTOPFlash reporter (20 ng) and TK-Renilla (5 ng) and the respective plasmid DNA as indicated using PromoFectin (PromoKine) transfection reagent according to the manufacturer’s protocol. Where indicated, cells were transfected with a SuperFOPFlash reporter instead of the SuperTOPFlash reporter. Each transfection was adjusted to 125 ng DNA/transfection with pcDNA3.1 empty vector. Where indicated, cells were transfected at 50–70% confluency with siRNA using the HiPerFect transfection reagent in 6 cm petri dishes according to the manufacturer’s protocol 24 h before seeding the cells for the reporter assays. Co-CM or Wnt3a-CM was added to HEK293 cells 24 h after DNA transfection. Where indicated, cells were treated with HA (400 μg/ml), hyaluronidase (HAse, 50 U/ml), blocking antibodies (3 μg/ml) or the chemical inhibitors (U0126, 15 μM; LY294002, 50 μM) 1 h prior to induction and over the whole induction experiment. Cells were lysed 48 h after DNA transfection with 1 × Passive Lysis Buffer (Promega), and the luciferase activity was measured using the Luminescence counter 1420 (PerkinElmer, Rodgau, Germany). TOPFlash experiments were normalized to co-transfected Renilla gene expression. Statistical analysis was performed using the Student’s t-test. A value of P<0.05 was considered statistically significant. In parallel to the reporter assay, transfected HEK293 cells were subjected to western blotting analysis.
β-Catenin and LRP6-activation assays
HeLa cells were seeded in six-well plates (1.5 × 105/well) and transfected as indicated with DNA (1 μg pcDNA3.1+500 ng CD44-Cyt or CD44-CytΔEz-BD) using PromoFectin (PromoKine) or with control- or CD44-siRNA as described above using HiPerFect (Qiagen). Cells were incubated for 2 h (LRP6-assay) or 3 h (β-catenin assay) with Co-CM or Wnt3a-CM and lysed 48 h after DNA transfection, respectively, 72 h after siRNA transfection and subjected to western blotting analysis.
Subcellular fractionation
To examine the subcellular localization of β-catenin, HeLa cells were seeded in 10 cm plates (5 × 105) and transfected with control- or CD44-siRNA. Seventy-two hours later, cells were treated for 6 h with Wnt3a-CM or Co-CM and subjected to subcellular fractionation as described in Huang et al.66 The membrane and cytosolic fraction were subjected to western blotting analysis using antibodies against CD44 (Hermes-3) and total β-catenin. Antibodies against Erk were used as control for the cytosolic fraction. To examine the membrane localization of rCD44v4-v7ΔCyt, HEK293 cells were transfected with rCD44v4-v7ΔCyt and 48 h later subjected to subcellular fractionation as described above. The membrane and the cytosolic fraction were subjected to western blotting analysis using an antibody against rCD44 (1.1ASML). Antibodies against transferrin receptor were used as control for the membrane fraction, and antibodies against Erk as control for the cytosolic fraction.
Co-immunoprecipitation and pulldown
For co-immunoprecipitation of endogenous proteins, HeLa cells were seeded in 10 cm plates (1 × 106) and incubated for the indicated time points with Wnt3a-CM. Cells were washed with ice cold PBS and lysed in lysis buffer (25 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% glycerol, 1% NP-40, 0.5 mM MgCl2, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM Na orthovanadate, and 1 mM aprotinine and leupeptin) for 30 min. Immunopreciptitation was performed on cleared lysates (12 000 r.p.m. for 15 min at 4 °C) with CD44 antibodies (Hermes-3) and protein G agarose beads (Merck) or mouse-IgG sepharose bead conjugates (IgG-control) (Cell Signaling) at 4 °C over night. The precipitates were washed in lysis buffer (3 × ) and boiled in SDS-sample buffer containing 100 mM DTT. Samples were subjected to western blotting analysis using anti-CD44 (Hermes-3) anti-LRP6 antibodies and stained using the ECL system. For co-immunoprecipitation of overexpressed FLAG-LRP6 and overexpressed rCD44 and rCD44ΔCyt constructs, cells were transfected in 10 cm plates with 2 μg FLAG-LRP6 together with 2 μg rCD44v4-v7 or 2 μg rCD44v4-v7ΔCyt plasmids using PromoFectin. Twenty-four hours after transfection, cells were treated for 30 min with Wnt3a-CM and subjected to co-immunoprecipitation as described above using an rCD44 antibody (1.1ASML) for the immunoprecipitation. Co-immunoprecipitated FLAG-LRP6 was detected using a FLAG-antibody. The co-immunoprecipitation of CD44 together with FLAG-LRP6 was performed similarly as described above using an antibody against FLAG-LRP6 for the immunoprecipitation. Co-immunoprecipitated CD44 was detected using an antibody against human CD44 (Hermes-3). For co-immunoprecipitation of overexpressed FLAG-LRP6 and endogenous CD44 in the presence of DN-Ezrin, cells were transfected in 10 cm plates with 2 μg FLAG-LRP6 together with either 2 μg empty vector (pCB6) or 1 μg DN-Ezrin and 1 μg empty vector using the PromoFectin transfection reagent. Twenty-four hours after transfection, cells were treated for 30 min with Wnt3a-CM and subjected to co-immunoprecipitation as described above using an hCD44 antibody (Hermes-3) for the immunoprecipitation. Co-immunoprecipitated FLAG-LRP6 was detected using a FLAG-antibody. For co-immunoprecipitation of overexpressed FLAG-LRP6 and overexpressed CD44s and CD44s-ERMmut, cells were transfected in 10 cm plates with 2 μg FLAG-LRP6 together with 2 μg CD44s or 2 μg CD44s-ERMmut plasmids using the PromoFectin transfection reagent. Twenty-four hours after transfection, cells were treated for 30 min with Wnt3a-CM and subjected to co-immunoprecipitation as described above using an CD44 antibody (Hermes-3) for the immunoprecipitation. Co-immunoprecipitated FLAG-LRP6 was detected using a FLAG-antibody. For co-immunoprecipitation of the cytoplasmic CD44-domain with a LRP6 mutant that lacks the extracellular domain (LRP6ΔE(1-4)), HeLa cells were seeded in 10 cm plates (1 × 106) and transfected with 2 μg CD44-Cyt-GST (described in Legge and Isacke37) together with 2 μg pEBG or 2 μg FLAG-LRP6ΔE(1-4). Forty-eight hours later, cells were washed with ice-cold PBS and lysed in lysis buffer (see above) for 30 min. The GST-pulldown was performed on cleared lysates (12 000 r.p.m. for 15 min at 4 °C) with Glutathione agarose beads (Sigma) at 4 °C over night. The precipitates were washed in lysis buffer (3 × ) boiled in SDS-sample buffer containing 100 mM DTT. Samples were subjected to western blotting analysis using anti-GST and anti-FLAG antibodies and stained using the ECL system.
Confocal microscopy
HeLa cells were seeded in 10 cm petri dishes and transfected at 50–70% confluency with control- or CD44pan-siRNA using HiPerFect (Qiagen) according to the manufacturer’s protocol. Twenty-four hours after siRNA transfection, cells were seeded on 20 mm glass cover slips (Menzel, Braunschweig, Germany) in 12-well plates. For β-catenin localization studies, cells were treated for 24 h either with Co-CM or Wnt3a-CM, washed once with PBS and fixed with methanol for 10 min at −20 °C. Afterwards cells were washed 3 × with PBS, blocked with 5% FBS in PBS for 1 h and incubated with a β-catenin antibody (D10A8,1:100) for 24 h at 4 °C. Subsequently, cells were washed 3 × with PBS, incubated with an donkey anti-rabbit-Alexa 546 antibody (0.4 μg/ml) for 2 h at room temperature, counterstained with DAPI for 30 min, washed another 3 × , mounted with Fluorescence Mounting Medium (Dako) and analyzed with an SP5 laser scanning confocal microscope (Leica, Wetzlar, Germany) with a 63 × objective. In parallel, transfected cells were subjected to western blotting analysis to confirm hCD44 knockdown. The percentage of cells with nuclear β-catenin was quantified and statistically analyzed using the double-sided Student’s t-test. A value of P<0.05 is considered statistically significant. For LRP6-GFP and Ror2-mCherry localization studies, cells were transfected with pcDNA3.1 empty vector (250 ng) or rCD44s (250 ng) or Mesd (50 ng) together with LRP6-GFP (250 ng), respectively, with pcDNA3.1 empty vector (250 ng) together with Ror2-mCherry (250 ng) using PromoFectin transfection reagent (PromoKine) according to the manufacturer’s protocol. Where indicated, HeLa cells seeded on 20 mm glass cover slips in 12-well plates were transfected with pcDNA3.1 empty vector, CD44-Cyt (250 ng), CD44-CytΔEzBD (250 ng) or DN-Ezrin (100 ng) together with LRP6-GFP (250 ng) constructs with PromoFectin. The total amount of plasmid DNA was adjusted to 1 μg with pcDNA3.1. Twenty-four hours after DNA transfection, cells were fixed with 4% paraformaldehyde for 10 min at room temperature (RT). Cells were incubated with DAPI (Life Technologies) and where indicated with an ER marker (ER-Tracker Red, Invitrogen) for 30 min at RT. Samples were mounted with Fluorescence Mounting Medium (Dako) and analyzed with an SP5 laser scanning confocal microscope with a 63 × objective. In parallel, transfected cells were subjected to western blotting analysis to confirm hCD44 knockdown and rCD44s overexpression. The membrane staining of LRP6-GFP and Ror2-mCherry was assessed by comparison of the GFP or mCherry channels with the corresponding brightfield channel. The ratio between cells with membrane or ER localized LRP6 to LRP6-GFP transfected cells was quantified and statistically analyzed using the Student’s t-test. A value of P<0.05 was considered statistically significant.
X. laevis studies
X. laevis embryos were in vitro fertilized, dejellied, cultured and injected as previously described.67 MOs (16 ng) were co-injected into one blastomere of X. laevis two-cell stage embryos with 4 pg dextrane-FITC as lineage tracer to identify the injected site at neurula stages and to sort left side injected embryos from right side injected embryos. Where indicated, xCD44-MO were co-injected with hCD44s (50 pg) or constitutive ABC (30 pg) expression vectors. Embryos were staged according to Nieuwkoop68 and fixed at stages 28–32 in MEMFA (0.1 M MOPS pH 7.2, 2 mM EGTA, 1 mM MgSO4 and 3.7% formaldehyde). In situ hybridization for tcf-4 and en-2 is described previously.31 Images were captured on a Leica MZFLIII microscope using a digital camera (Qimaging, Surrey, BC, Canada) and the Improvision software (PerkinElmer, Rodgau, Germany). Statistical analysis was performed using the Student’s t-test. A value of P<0.05 was considered statistically significant.
Statistical analysis
The Student’s t-test was used for comparison of two samples. Calculation of the average was performed using at least three biological replicates. P-values<0.05 were considered significant. Error bars indicate S.D.
Abbreviations
- ABC:
-
active β-catenin
- APC:
-
adenomatous polyposis coli
- Arp2/3:
-
actin-related proteins 2/3
- CD44:
-
cluster of differentiation 44
- Dkk1:
-
Dickkopf1
- DN:
-
dominant negative
- Dsh:
-
dishevelled
- En-2:
-
engrailed-2
- ER:
-
endoplasmic reticulum
- Erk:
-
Extracellular-signal regulated kinases
- ERM:
-
Ezrin-Radixin-Moesin
- Ez:
-
Ezrin
- GFP:
-
green fluorescent protein
- GST:
-
glutathione S-transferase
- HA:
-
hyaluronan
- ICD:
-
intracellular domain
- Lgr5:
-
Leucine-rich repeat-containing G-protein coupled receptor 5
- LEF:
-
lymphoid enhancer factor
- LRP:
-
low-density lipoprotein receptor-related protein
- MAPK:
-
mitogen-activated protein kinase
- Mesd:
-
mesoderm development
- MO:
-
morpholino
- N-WASP:
-
neural Wiskott-Aldrich syndrome protein
- PI3K:
-
phosphoinositide 3-kinase
- Ror2:
-
receptor tyrosine kinase-like orphan receptor 2
- Ser1490:
-
Serine 1490
- TCF:
-
T-cell factor
References
Logan CY, Nusse R . The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 2004; 20: 781–810.
Clevers H . Wnt/beta-catenin signaling in development and disease. Cell 2006; 127: 469–480.
Clevers H, Nusse R . Wnt/beta-catenin signaling and disease. Cell 2012; 149: 1192–1205.
He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT et al. Identification of c-MYC as a target of the APC pathway. Science 1998; 4 281: 1509–1512.
Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F . Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol 2002; 22: 1172–1183.
Yan D, Wiesmann M, Rohan M, Chan V, Jefferson AB, Guo L et al. Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/beta -catenin signaling is activated in human colon tumors. Proc Natl Acad Sci USA 2001; 98: 14973–14978.
Wielenga VJ, Smits R, Korinek V, Smit L, Kielman M, Fodde R et al. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am J Pathol 1999; 154: 515–523.
Naor D, Sionov RV, Ish-Shalom D . CD44: structure, function and association with the malignant process In: Vande Woude GF, Klein G (eds). Advances in Cancer Research. Elsevier: Amsterdam, Netherlands, 1997, pp 243–318.
Ponta H, Sherman L, Herrlich PA . CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003; 4: 33–45.
Orian-Rousseau V . CD44, a therapeutic target for metastasising tumours. Eur J Cancer 2010; 46: 1271–1277.
Fuchs K, Hippe A, Schmaus A, Homey B, Sleeman JP, Orian-Rousseau V . Opposing effects of high- and low-molecular weight hyaluronan on CXCL12-induced CXCR4 signaling depend on CD44. Cell Death Dis 2013; 4: e819.
Kuhnert F, Davis CR, Wang HT, Chu P, Lee M, Yuan J et al. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci USA 2004; 101: 266–271.
Zeilstra J, Joosten SP, van Andel H, Tolg C, Berns A, Snoek M et al. Stem cell CD44v isoforms promote intestinal cancer formation in Apc(min) mice downstream of Wnt signaling. Oncogene 2014; 33: 665–670.
Roca X, Mate JL, Ariza A, Munoz-Marmol AM, von Uexkull-Guldeband C, Pellicer I et al. CD44 isoform expression follows two alternative splicing pathways in breast tissue. Am J Pathol 1998; 153: 183–190.
Gansauge F, Gansauge S, Zobywalski A, Scharnweber C, Link KH, Nussler AK et al. Differential expression of CD44 splice variants in human pancreatic adenocarcinoma and in normal pancreas. Cancer Res 1995; 55: 5499–5503.
Fonseca I, Moura Nunes JF, Soares J . Expression of CD44 isoforms in normal salivary gland tissue: an immunohistochemical and ultrastructural study. Histochem Cell Biol 2000; 114: 483–488.
Shimabukuro K, Toyama-Sorimachi N, Ozaki Y, Goi T, Furukawa K, Miyasaka M et al. The expression patterns of standard and variant CD44 molecules in normal uterine cervix and cervical cancer. Gynecol Oncol 1997; 64: 26–34.
Kasper M, Gunthert U, Dall P, Kayser K, Schuh D, Haroske G et al. Distinct expression patterns of CD44 isoforms during human lung development and in pulmonary fibrosis. Am J Respir Cell Mol Biol 1995; 13: 648–656.
Mackay CR, Terpe H-J, Stauder R, Marston WL, Stark H, Günthert U . Expression and modulation of CD44 variant isoforms in humans. J Cell Biol 1994; 124: 71–82.
Wielenga VJ, Heider KH, Offerhaus GJ, Adolf GR, van den Berg FM, Ponta H et al. Expression of CD44 variant proteins in human colorectal cancer is related to tumor progression. Cancer Res 1993; 53: 4754–4756.
Bloor BK, Jelvagharan M, White KN, Odell EW . Characterization of CD44 splicing patterns in normal keratinocytes, dysplastic and squamous carcinoma cell lines. Int J Oncol 2001; 18: 1053–1059.
Herold-Mende C, Seiter S, Born AI, Patzelt E, Schupp M, Zoller J et al. Expression of CD44 splice variants in squamous epithelia and squamous cell carcinomas of the head and neck. J Pathol 1996; 179: 66–73.
Yu Q, Toole BP . Common pattern of CD44 isoforms is expressed in morphogenetically active epithelia. Dev Dyn 1997; 208: 1–10.
Wielenga VJ, van der Neut R, Offerhaus GJ, Pals ST . CD44 glycoproteins in colorectal cancer: expression, function, and prognostic value. Adv Cancer Res 2000; 77: 169–187.
Zeilstra J, Joosten SP, Dokter M, Verwiel E, Spaargaren M, Pals ST . Deletion of the WNT target and cancer stem cell marker CD44 in Apc(Min/+) mice attenuates intestinal tumorigenesis. Cancer Res 2008; 68: 3655–3661.
Misra S, Hascall VC, De Giovanni C, Markwald RR, Ghatak S . Delivery of CD44shRNA/nanoparticles within cancer cells: perturbation of hyaluronan/CD44v6 interactions and reduction in adenoma growth in Apc Min/+ mice. J Biol Chem 2009; 284: 12432–12446.
Chang G, Zhang H, Wang J, Zhang Y, Xu H, Wang C et al. CD44 targets Wnt/beta-catenin pathway to mediate the proliferation of K562 cells. Cancer Cell Int 2013; 13: 117.
Misra S, Obeid LM, Hannun YA, Minamisawa S, Berger FG, Markwald RR et al. Hyaluronan constitutively regulates activation of COX-2-mediated cell survival activity in intestinal epithelial and colon carcinoma cells. J Biol Chem 2008; 283: 14335–44.
Krejci P, Aklian A, Kaucka M, Sevcikova E, Prochazkova J, Masek JK et al. Receptor tyrosine kinases activate canonical WNT/beta-catenin signaling via MAP kinase/LRP6 pathway and direct beta-catenin phosphorylation. PLoS One 2012; 7: e35826.
Bourguignon LY, Peyrollier K, Gilad E, Brightman A . Hyaluronan-CD44 interaction with neural Wiskott-Aldrich syndrome protein (N-WASP) promotes actin polymerization and ErbB2 activation leading to beta-catenin nuclear translocation, transcriptional up-regulation, and cell migration in ovarian tumor cells. J Biol Chem 2007; 282: 1265–1280.
Koenig SF, Brentle S, Hamdi K, Fichtner D, Wedlich D, En2 Gradl D . Pax2/5 and Tcf-4 transcription factors cooperate in patterning the Xenopus brain. Dev Biol 2010; 340: 318–328.
McGrew LL, Takemaru K, Bates R, Moon RT . Direct regulation of the Xenopus engrailed-2 promoter by the Wnt signaling pathway, and a molecular screen for Wnt-responsive genes, confirm a role for Wnt signaling during neural patterning in Xenopus. Mech Dev 1999; 87: 21–32.
König H, Moll J, Ponta H, Herrlich P . Trans-acting factors regulate the expression of CD44 splice variants. EMBO J 1996; 15: 4030–4039.
Cheng C, Yaffe MB, Sharp PA . A positive feedback loop couples Ras activation and CD44 alternative splicing. Genes Dev 2006; 20: 1715–1720.
Radtke F, Clevers H . Self-renewal and cancer of the gut: two sides of a coin. Science 2005; 307: 1904–1909.
Peach RJ, Hollenbaugh D, Stamenkovic I, Aruffo A . Identification of hyaluronic acid binding sites in the extracellular domain of CD44. J Cell Biol 1993; 122: 257–264.
Legg JW, Isacke CM . Identification and functional analysis of the ezrin-binding site in the hyaluronan receptor, CD44. Curr Biol 1998; 8: 705–708.
Orian-Rousseau V, Chen L, Sleeman JP, Herrlich P, Ponta H . CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev 2002; 16: 3074–3086.
Algrain M, Turunen O, Vaheri A, Louvard D, Arpin M . Ezrin contains cytoskeleton and membrane binding domains accounting for its proposed role as a membrane-cytoskeletal linker. J Cell Biol 1993; 120: 129–139.
Davidson G, Wu W, Shen J, Bilic J, Fenger U, Stannek P et al. Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature 2005; 438: 867–872.
Tamai K, Zeng X, Liu C, Zhang X, Harada Y, Chang Z et al. A mechanism for Wnt coreceptor activation. Mol Cell 2004; 13: 149–156.
Hsieh JC, Lee L, Zhang L, Wefer S, Brown K, DeRossi C et al. Mesd encodes an LRP5/6 chaperone essential for specification of mouse embryonic polarity. Cell 2003; 112: 355–367.
Ori M, Nardini M, Casini P, Perris R, Nardi I . XHas2 activity is required during somitogenesis and precursor cell migration in Xenopus development. Development 2006; 133: 631–640.
Kunz M, Herrmann M, Wedlich D, Gradl D . Autoregulation of canonical Wnt signaling controls midbrain development. Dev Biol 2004; 273: 390–401.
Hidalgo-Sanchez M, Millet S, Bloch-Gallego E, Alvarado-Mallart RM . Specification of the meso-isthmo-cerebellar region: the Otx2/Gbx2 boundary. Brain Res Brain Res Rev 2005; 49: 134–149.
Wehrli M, Dougan ST, Caldwell K, O'Keefe L, Schwartz S, Vaizel-Ohayon D et al. arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature 2000; 407: 527–530.
Bilic J, Huang YL, Davidson G, Zimmermann T, Cruciat CM, Bienz M et al. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science 2007; 316: 1619–1622.
Galli C, Piemontese M, Lumetti S, Ravanetti F, Macaluso GM, Passeri G . Actin cytoskeleton controls activation of Wnt/beta-catenin signaling in mesenchymal cells on implant surfaces with different topographies. Acta Biomater 2012; 8: 2963–2968.
Marion S, Hoffmann E, Holzer D, Le Clainche C, Martin M, Sachse M et al. Ezrin promotes actin assembly at the phagosome membrane and regulates phago-lysosomal fusion. Traffic 2011; 12: 421–437.
Deretic D, Traverso V, Parkins N, Jackson F, Rodriguez de Turco EB, Ransom N . Phosphoinositides, ezrin/moesin, and rac1 regulate fusion of rhodopsin transport carriers in retinal photoreceptors. Mol Biol Cell 2004; 15: 359–370.
Orian-Rousseau V, Morrison H, Matzke A, Kastilan T, Pace G, Herrlich P et al. Hepatocyte growth factor-induced Ras activation requires ERM proteins linked to both CD44v6 and F-actin. Mol Biol Cell 2007; 18: 76–83.
Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G et al. Two members of the Tcf family implicated in Wnt/beta-catenin signaling during embryogenesis in the mouse. Mol Cell Biol 1998; 18: 1248–1256.
Pinto D, Gregorieff A, Begthel H, Clevers H . Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev 2003; 17: 1709–1713.
Shibamoto S, Higano K, Takada R, Ito F, Takeichi M, Takada S . Cytoskeletal reorganization by soluble Wnt-3a protein signalling. Genes Cells 1998; 3: 659–670.
Matzku S, Wenzel A, Liu S, Zoller M . Antigenic differences between metastatic and nonmetastatic BSp73 rat tumor variants characterized by monoclonal antibodies. Cancer Res 1989; 49: 1294–1299.
Sleeman JP, Arming S, Moll JF, Hekele A, Rudy W, Sherman LS et al. Hyaluronate-independent metastatic behavior of CD44 variant-expressing pancreatic carcinoma cells. Cancer Res 1996; 56: 3134–3141.
Avigdor A, Goichberg P, Shivtiel S, Dar A, Peled A, Samira S et al. CD44 and hyaluronic acid cooperate with SDF-1 in the trafficking of human CD34+ stem/progenitor cells to bone marrow. Blood 2004; 103: 2981–2989.
Matzke A, Herrlich P, Ponta H, Orian-Rousseau V . A five-amino-acid peptide blocks Met- and Ron-dependent cell migration. Cancer Res 2005; 65: 6105–6110.
Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM et al. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature 2002; 417: 664–667.
Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y et al. LDL-receptor-related proteins in Wnt signal transduction. Nature 2000; 407: 530–535.
Cruciat CM, Dolde C, de Groot RE, Ohkawara B, Reinhard C, Korswagen HC et al. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-beta-catenin signaling. Science 2013; 339: 1436–1441.
Aberle H, Bauer A, Stappert J, Kispert A, Kemler R . beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J 1997; 16: 3797–3804.
Feike AC, Rachor K, Gentzel M, Schambony A . Wnt5a/Ror2-induced upregulation of xPAPC requires xShcA. Biochem Biophys Res Commun 2010; 400: 500–506.
Landesman Y, Sokol SY . Xwnt-2b is a novel axis-inducing Xenopus Wnt, which is expressed in embryonic brain. Mech Dev 1997; 63: 199–209.
Konig A, Gradl D, Kuhl M, Wedlich D . The HMG-box transcription factor XTcf-4 demarcates the forebrain-midbrain boundary. Mech Dev 2000; 93: 211–214.
Huang Z, Cheng Y, Chiu PM, Cheung FM, Nicholls JM, Kwong DL et al. Tumor suppressor Alpha B-crystallin (CRYAB) associates with the cadherin/catenin adherens junction and impairs NPC progression-associated properties. Oncogene 2012; 31: 3709–3720.
Peng HB . Xenopus laevis: practical uses in cell and molecular biology. Solutions and protocols. Methods Cell Biol 1991; 36: 657–662.
Nieuwkoop PD . The ‘organization centre’. 3. Segregation and pattern formation in morphogenetic fields. Acta Biotheor 1967; 17: 178–194.
Acknowledgements
This work was supported by grants from the Deutsche Forschungsgemeinschaft (OR124/12-1, to VOR and MS) and FOR1036 (to GD and DG).
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by E Wagner
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
Schmitt, M., Metzger, M., Gradl, D. et al. CD44 functions in Wnt signaling by regulating LRP6 localization and activation. Cell Death Differ 22, 677–689 (2015). https://doi.org/10.1038/cdd.2014.156
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2014.156
This article is cited by
-
Crosstalk between colorectal CSCs and immune cells in tumorigenesis, and strategies for targeting colorectal CSCs
Experimental Hematology & Oncology (2024)
-
Integrated whole transcriptome profiling revealed a convoluted circular RNA-based competing endogenous RNAs regulatory network in colorectal cancer
Scientific Reports (2024)
-
Loss of Epidermal Homeostasis Underlies the Development of Squamous Cell Carcinoma
Stem Cell Reviews and Reports (2023)
-
Over Expression of Cancer Stem Cell Marker CD44 and Its Clinical Significance in Patients with Oral Squamous Cell Carcinoma
Indian Journal of Otolaryngology and Head & Neck Surgery (2023)
-
Exosomes: the key of sophisticated cell–cell communication and targeted metastasis in pancreatic cancer
Cell Communication and Signaling (2022)