Abstract
Neuroblastoma is an embryonal malignancy of the sympathetic nervous system. Spontaneous regression and differentiation of neuroblastoma is observed in a subset of patients, and has been suggested to represent delayed activation of physiologic molecular programs of fetal neuroblasts. Homeobox genes constitute an important family of transcription factors, which play a fundamental role in morphogenesis and cell differentiation during embryogenesis. In this study, we demonstrate that expression of the majority of the human HOX class I homeobox genes is significantly associated with clinical covariates in neuroblastoma using microarray expression data of 649 primary tumors. Moreover, a HOX gene expression-based classifier predicted neuroblastoma patient outcome independently of age, stage and MYCN amplification status. Among all HOX genes, HOXC9 expression was most prominently associated with favorable prognostic markers. Most notably, elevated HOXC9 expression was significantly associated with spontaneous regression in infant neuroblastoma. Re-expression of HOXC9 in three neuroblastoma cell lines led to a significant reduction in cell viability, and abrogated tumor growth almost completely in neuroblastoma xenografts. Neuroblastoma growth arrest was related to the induction of programmed cell death, as indicated by an increase in the sub-G1 fraction and translocation of phosphatidylserine to the outer membrane. Programmed cell death was associated with the release of cytochrome c from the mitochondria into the cytosol and activation of the intrinsic cascade of caspases, indicating that HOXC9 re-expression triggers the intrinsic apoptotic pathway. Collectively, our results show a strong prognostic impact of HOX gene expression in neuroblastoma, and may point towards a role of Hox-C9 in neuroblastoma spontaneous regression.
Similar content being viewed by others
Main
Class I homeobox (Hox) transcription factors constitute an important family of developmental regulators, which play a fundamental role in morphogenesis and cell differentiation during embryogenesis.1 In neural progenitors, HOX gene expression is controlled by retinoic acid (RA), fibroblast growth factors and wingless-type family members.2, 3 A deregulated expression of HOX genes has been observed in a number of malignancies.4, 5, 6, 7
Neuroblastoma, an embryonal tumor of the sympathetic nervous system, originates from primordial neural crest cells, which are destined for sympathetic differentiation. This pediatric solid tumor shows remarkable variations in clinical presentations ranging from aggressive, therapy-resistant progression to spontaneous regression, which regularly occurs in infants both with localized and metastasized disease. Furthermore, neuroblastoma cells show the potential to differentiate toward a sympathetic ganglion cell phenotype.8 It has thus been suggested that the physiologic molecular program of neuroblast differentiation and growth control is disrupted in neuroblastoma.9 This developmental arrest may be reversible in spontaneously regressing neuroblastoma, in which a delayed activation of naturally occurring processes of programmed cell death has been suspected.10, 11, 12 According to its embryonic nature, many genes aberrantly expressed in neuroblastoma are involved in developmental processes.13, 14 Among others, several transcription factors involved in the development of autonomic neural crest derivates, such as MYCN and PHOX2B, are implicated in the pathogenesis of neuroblastoma.15, 16, 17, 18 Furthermore, various HOX genes have been described to be aberrantly expressed in neuroblastoma cell lines and primary tumors.19, 20 In a recent study, Mao et al.21 reported that HOXC9 expression is decreased in advanced-stage neuroblastoma and is involved in cell cycle control and the processes of neuroblastoma cell differentiation.
In this study, we aimed at determining the association of class I HOX gene expression patterns with prognostic markers and outcome in neuroblastoma. Because HOXC9 was not only associated with favorable outcome but also with spontaneous regression, we investigated the functional consequences of HOXC9 re-expression on neuroblastoma growth and programmed cell death.
Results
Expression of the HOX gene cluster is deregulated in neuroblastoma
The expression patterns of the 39 class I HOX genes were analyzed in 649 neuroblastoma samples by microarrays, and the association with prognostic markers and patient outcome was determined. The expression of the majority of HOX genes correlated significantly with clinical covariates in neuroblastoma. Elevated expression of HOXD genes, particularly HOXD3, HOXD8, HOXD9 and HOXD10, was predominantly associated with unfavorable prognostic markers and poor outcome (Supplementary Table S1 and Supplementary Figures S1 and S2). Similarly, we observed increased expression levels of more posterior HOX genes (HOXA10, HOXA11, HOXA13, HOXC12 and HOXC13) in neuroblastomas with unfavorable characteristics (Supplementary Table S1). In contrast, elevated expression of the majority of the remaining HOX genes was significantly correlated with favorable prognostic markers (Supplementary Table S1).
Prediction of neuroblastoma outcome based on a HOX gene expression signature
To assess the impact of HOX gene expression on neuroblastoma outcome, we developed a HOX gene expression-based classifier (Table 1a and Supplementary Table S2) using a training cohort of 75 neuroblastoma patients with maximal divergent outcome. In the training set, the classification accuracy was 85% as assessed by cross-validation. In a validation subset of 215 patients who matched the outcome criteria of the training set, the 33 HOX gene signature predicted patient outcome with an accuracy of 76% (Table 1b). In the entire validation set (n=574), the classifier accurately discriminated patients with favorable and unfavorable outcome (favorable: n=265; 5-year event-free survival (EFS), 80.7±2.6%; 5-year overall survival (OS), 96.0±1.2%; unfavorable: n=309; 5-year EFS, 49.5±3.0%; 5-year OS, 67.1±2.8%; both P<0.001). In multivariate Cox regression models based on EFS and OS, the HOX classifier predicted patient outcome independently of age, stage and MYCN amplification status (Table 1c).
Elevated HOXC9 expression is associated with favorable prognostic markers, beneficial patient outcome and spontaneous regression
Elevated HOXC9 expression levels strongly correlated with lower stages, age <18 months at diagnosis, lack of MYCN amplification, lack of 1p loss and favorable gene expression-based classification22, 23 (Figure 1a). Kaplan–Meier estimates were calculated by using two independent neuroblastoma patient cohorts. The first cohort consisted of 244 neuroblastoma patients, which has been published previously by our group22 and has been used by Mao and co-workers21 for clinical analysis of HOXC9 expression. This cohort was used to determine cutoff values of HOXC9 expression levels by quartiles. The cutoff values were applied on an independent set of 405 neuroblastoma patients to determine clinical courses of patients with high, intermediate–high, intermediate–low and low HOXC9 expressions. Patients with high HOXC9 expression had a significantly better outcome with a 5-year OS of 95±2% as compared to patients with low HOXC9 expression (5-year OS, 56±5%; Figure 1b). Likewise, HOXC9 expression levels strongly correlated with improved EFS (high HOXC9 expression, 5-year EFS, 82±4% versus low HOXC9 expression, 5-year EFS, 37±5%; Figure 1c). In addition, multivariate Cox regression models based on EFS and OS, considering established risk markers (MYCN status, tumor stage and patient age at diagnosis), determined HOXC9 expression as a significant independent prognostic marker for both EFS and OS (Table 2). We also investigated HOXC9 expression levels in infant neuroblastoma that had regressed without any chemotherapy, and compared these with infant stage 4 neuroblastoma. Notably, HOXC9 transcript levels were significantly higher in neuroblastoma showing spontaneous regression (Figure 1d and Supplementary Table S3). Taken together, these results demonstrate that HOXC9 transcript levels discriminate neuroblastoma patients with favorable and unfavorable outcome and indicate that elevated HOXC9 expression is associated with spontaneous regression.
(a) Association of HOXC9 expression with prognostic markers in 649 neuroblastomas as determined by microarray analysis. PAM classifier, gene expression-based classifier as defined in the main text. Boxes, median expression values (horizontal line) and 25th and 75th percentiles; whiskers, distances from the end of the box to the largest and smallest observed values that are <1.5 box lengths from either end of the box; open circles, outlying values; asterisks, extreme outlying values. (b) OS probability in the validation set of neuroblastoma patients according to HOXC9 expression. (c) EFS probability in the validation set of neuroblastoma patients according to HOXC9 expression. (d) HOXC9 expression levels in spontaneously regressing and stage 4 infant neuroblastomas. (e) HOXC9 expression according to primary tumor localization. Amp, amplification; norm, normal; aberr; aberration; fav, favorable; unfav, unfavorable; SR, spontaneously regressive; NK, neck; C, chest; ABD, abdomen; AG, adrenal gland; Loc, localization
Elevated HOXC9 expression is associated with abdominal neuroblastoma
As the expression of HOX genes is not only temporally but also spatially regulated during embryonic development, HOXC9 expression were compared in neuroblastoma according to the primary tumor site. We observed significantly higher HOXC9 expression levels in tumors with abdominal location in comparison to neuroblastomas located at the neck/chest or at adrenal glands (Figure 1e), which is in line with the embryonal expression pattern of HOX genes: HOX genes located towards the 5′end of the HOX cluster, such as HOXC9, are expressed more posterior in the body during development. Analogous expression patterns, however, were observed for only few of the remaining 5′-located HOX genes (Supplementary Figure S3a). Interestingly, the spatial expression pattern of HOXC9 might not have been expected from the association of primary tumor sites with patient outcome.24, 25 In line with these reports, we observed a better outcome of patients with neck/chest neuroblastoma in comparison to tumors located at the adrenal glands or in the abdomen (neck/chest versus adrenal glands versus abdomen, 5-year EFS, 84±5% versus 69±3% versus 63±4%, respectively; 5-year OS, 97±2% versus 83±2% versus 85±3%, respectively; Supplementary Figures S3b and S3c).
Numerical gain of chromosome 12 correlates with elevated expression of HOXC9
We next aimed to evaluate whether genetic or epigenetic aberrations of the HOXC9 locus are associated with deregulated HOXC9 expression. First, we analyzed array-comparative genomic hybridization (aCGH) profiles of 209 neuroblastoma samples and compared the results with corresponding microarray gene expression data of the same tumors. Genomic aberrations of chromosome 12 were detected in 92/209 (44.0%) of the samples (Figure 2a and Supplementary Table S4). We most frequently observed numerical gains of the entire chromosome (n=64, 30.6%). Numerical chromosome 12 loss occurred only in four cases. Segmental alterations of chromosome 12 were detected in 24 tumors (11.5%), most of which were segmental gains not affecting the HOXC9 locus (n=18, 8.6%). Analysis of HOXC9 gene expression data revealed significantly higher transcript levels in tumors with numerical chromosome 12 gains in comparison to tumors in which the HOXC9 locus was not affected by genomic alterations (Figure 2b).
To examine whether inactivating mutations might contribute to diminished HOXC9 expression in neuroblastoma, we sequenced the genomic HOXC9 locus of 46 primary tumors with low HOXC9 expression. A total of 16 unique sequence variants were detected (Supplementary Table S5). Eight of these represented known SNPs, whereas the remaining sequence variants were novel. The latter variants affected either non-coding sequences or were synonymous, and occurred infrequently in our cohort. Taken together, these data suggest that mutations in the HOXC9 locus are not a major cause of reduced HOXC9 expression levels in unfavorable neuroblastoma.
To determine whether epigenetic regulation might contribute to differences in HOXC9 expression in neuroblastoma, we analyzed the methylation status of 26 CpG sites located in the HOXC9 promoter region. DNA samples from 46 neuroblastoma tumors with differing HOXC9 expression levels (high and low HOXC9 expression as defined in Materials and Methods, n=23 per subgroup) and 2 neuroblastoma cell lines (SK-N-AS and IMR-32) with loss of HOXC9 expression were analyzed. Unsupervised one-way hierarchical clustering of CpG site methylation revealed a largely homogeneous methylation pattern in tumors of both subgroups and cell lines (Supplementary Figure S4b), suggesting that downregulation of HOXC9 does not result from altered CpG methylation patterns.
Doxycycline-inducible expression of Hox-C9 in neuroblastoma cell lines
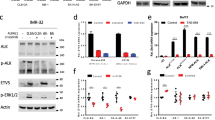
HOXC9 expression was restored in three neuroblastoma cell lines. Polyclonal Hox-C9-expressing neuroblastoma cells were compared with polyclonal GFP-expressing cells (Figure 3a) due to promoter leakage of the pRevTRE Vector System (Figure 3b). Recombinant Hox-C9 protein levels were comparable to physiological protein levels observed in neuroblastoma patients with high Hox-C9 expression (Figure 3c).
Inducible expression of Hox-C9 in SK-N-AS, IMR-32 and CHP-212 cells and physiological Hox-C9 levels in primary neuroblastomas as determined by western blot analysis. (a) Inducible expression of Hox-C9- versus GFP-expressing control cells. (b) Time course of Hox-C9 expression. (c) Representative neuroblastoma tumor samples with high and low Hox-C9 expression. Dox, doxycycline; NB, neuroblastoma
HOXC9 expression inhibits growth of neuroblastoma cells in vitro and in vivo
To investigate whether HOXC9 expression affects growth properties in neuroblastoma cells, polyclonal HOXC9-expressing IMR-32, SK-N-AS and CHP-212 cells were compared with polyclonal GFP-expressing controls. Cell proliferation was assayed for up to 8 days using Trypan blue dye exclusion tests. In all cell lines, the number of viable cells at day 8 was significantly lower compared with GFP-induced controls (Figure 4a). To determine whether reduced proliferation upon HOXC9 re-expression might be due to impaired cell cycle progression, the DNA content of HOXC9-expressing cells was assessed by flow cytometry. We observed a significant increase of the G0/G1 peak at day 7 after Hox-C9 induction in all three cell lines (Figure 4b). To investigate the influence of HOXC9 on anchorage-independent clonal growth, we preformed soft agar assays. A marked reduction in colony formation was observed in both SK-N-AS and IMR-32 cells in comparison to control cells (Figure 4c). Two xenograft mouse model systems were used to assess the effect of Hox-C9 on neuroblastoma tumor growth in vivo. GFP-expressing SK-N-AS and IMR-32 control cells formed rapidly growing tumors with mean tumor volumes of 1905±295 mm3 at day 17 and 1323±404 mm3 at day 22, respectively. In contrast, tumor growth was almost completely abrogated in mice inoculated with HOXC9-expressing SK-N-AS and IMR-32 (tumor volumes of 79±31 mm3 at day 17 and 173±129 mm3 at day 22, respectively; Figure 4d).
HOXC9 re-expression inhibits growth of neuroblastoma cells in vitro and in vivo. Hox-C9-induced changes in (a) cell proliferation (Trypan blue dye exclusion analysis), (b) cell cycle distribution (fluorescence-activated cell sorting (FACS)) and (c) soft agar colony formation. (d) Hox-C9 expression impede tumor growth in SK-N-AS and IMR-32 neuroblastoma xenografts (n=8 mice per group). Error bars indicate S.D.
Hox-C9-induced neuronal differentiation of IMR-32 cells is accompanied by the downregulation of REST
Morphological signs of neuronal differentiation were assessed by microscopic examination in all three cell lines after HOXC9 re-expression. In IMR-32 cells, HOXC9 re-expression led to a neuronal-like phenotype with a large network of cells interconnected by long neurite elongations (Figure 5a), while similar changes were not observed in the other cell lines. To determine whether this morphological alteration induced by Hox-C9 was accompanied by the upregulation of genes involved in neuronal differentiation, we analyzed expression levels of the neuron-related markers DNER, NTS, PTN, NNAT, TRKA and NEFL by quantitative real-time reverse transcriptase-polymerase chain reactions (qRT-PCR) and microarray analysis. In line with the morphological changes, all markers were upregulated upon HOXC9 re-expression (Figures 5b and c). A similar trend was observed for additional markers associated with neuronal differentiation (Supplementary Figure S5a and S5b). At the same time, the RE1-silencing transcription factor REST, a master negative regular of neurogenesis, was downregulated in Hox-C9-induced IMR-32 cells (Figures 5d and e).
Hox-C9 induces neuronal differentiation in IMR-32 cells. (a) Morphological changes of IMR-32 cells upon Hox-C9 and GFP induction at day 21. HOXC9 expression upregulates neuron-related markers in IMR-32 cells as determined by (b) qRT-PCR and (c) oligonucleotide microarrays. Hox-C9 expression downregulates REST as determined by (d) oligonucleotide microarrays and (e) qRT-PCR. Error bars indicate S.D.
HOXC9 re-expression induces apoptosis in neuroblastoma cells
We next examined whether apoptosis contributes to the inhibition of neuroblastoma cell growth after HOXC9 re-expression. First, externalization of phosphatidylserine was analyzed by flow cytometry using Annexin-V staining. We observed a significant increase of the Annexin-V-binding fraction in SK-N-AS and CHP-212 cells 168 h after HOXC9 re-expression in comparison to control cells (Figure 6a). Second, DNA fragmentation was assessed using the terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay in SK-N-AS, IMR-32 and CHP-212 cells. HOXC9-expressing neuroblastoma cells showed a significantly higher fraction of apoptotic cells as compared with GFP-expressing controls (Figure 6b). These results were supported by FACS analysis following Hox-C9 induction in SK-N-AS, IMR-32 and CHP-212 cells, in which Nicoletti labeling and assessment of the fraction of sub-G1 events indicated an accumulation of cells with fragmented DNA. In all investigated cell lines, the number of apoptotic cells was significantly increased by HOXC9 re-expression as compared with GFP-induced controls (Figure 6c). Taken together, these findings indicate that Hox-C9 affects growth properties in human neuroblastoma cells not only by cell cycle regulation but also by induction of apoptosis.
Hox-C9 induces apoptosis in neuroblastoma cell lines by activating the intrinsic apoptotic pathway. (a) Proportion of Annexin-V-positive cells as determined by fluorescence-activated cell sorting (FACS). (b) Proportion of cells showing apoptotic nuclei as determined by fluorescence microscopy. (c) Proportion of cells with subnormal DNA content as determined by FACS. (d) Protein expression of Bcl-2, cytosolic cytochrome c and cleaved caspase-9, caspase-7 and caspase-3 upon Hox-C9 and GFP induction as determined by western blot analysis (for densitometric quantification see Supplementary Figure S6). (e) BCL-2 and BAX expression in IMR-32 and SK-N-AS cells following Hox-C9 induction as determined by microarrays. (f) BCL-2 expression in IMR-32 cells, and BAX expression in SK-N-AS and CHP-212 cells following Hox-C9 induction as determined by qRT-PCR. Error bars indicate S.D. Dox, doxycycline
Hox-C9 activates the intrinsic pathway of apoptosis
To further analyze Hox-C9-induced cell death, the expression of pro- and antiapoptotic regulators was examined by immunoblot, qRT-PCR and microarray analysis. Apoptotic cell death was associated with a strong increase of cytochrome c in the cytosolic fraction of Hox-C9-induced cells (Figure 6d), suggesting a loss of mitochondrial membrane potential and involvement of mitochondria in Hox-C9-induced cell death. In IMR-32 cells, we observed downregulation of BCL-2 both at the transcript level and the protein level, while the expression levels remained unchanged in SK-N-AS and CHP-212 cells (Figures 6d–f). In the latter cell lines, however, we observed a marked upregulation of BAX mRNA expression upon induction of Hox-C9 (Figures 6e and f). These data suggest that apoptosis induced by HOXC9 re-expression may be conferred by a shifted BCL-2/BAX ratio as a consequence of either increased expression of proapoptotic BAX or decreased expression of antiapoptotic Bcl-2. Subsequently, we investigated whether procaspase-9, the initial caspase in the mitochondrial apoptotic cascade, is activated upon cytochrome c release. We observed an increase of cleaved 35 kDa fragments of activated caspase-9 in all cell lines upon HOXC9 re-expression (Figure 6d). In addition, a 37 kDa cleaved fragment of caspase-9 was observed in SK-N-AS and CHP-212 cells (Figure 6d), which is indicative of a feedback amplification loop induced by activated caspase-3.26 Finally, we observed an increased activation of the downstream effector caspase-3 and -7 in all three cell lines (Figure 6d) upon HOXC9 re-expression. Taken together, these data indicate that HOXC9 expression can activate the intrinsic pathway of apoptosis and thereby trigger neuroblastoma cell death.
Hox-C9 affects transcriptional pathways regulating differentiation and cell death
To gain further insights into the molecular processes occurring upon HOXC9 re-expression, we analyzed gene expression profiles of IMR-32 and SK-N-AS cells after Hox-C9 induction using microarrays. We used gene ontology (GO) annotations to find classes of genes that are significantly over-represented in gene sets that were either up- or downregulated after HOXC9 re-expression (Supplementary Table S6). Many GO categories significantly enriched for genes upregulated in HOXC9-expressing IMR-32 cells were related to neuronal functions and differentiation, thereby reflecting the Hox-C9-associated differentiation phenotype in IMR-32 cells (Supplementary Table S6 and Supplementary Figure S7a). In SK-N-AS cells, we observed GO categories significantly enriched for genes upregulated by Hox-C9 that were associated with cell death (Supplementary Table S6 and Supplementary Figure S7c). These results are well in line with the observed phenotypes of these two neuroblastoma cell lines after HOXC9 re-expression: While growth inhibition mediated by apoptosis is the predominant characteristic of HOXC9-expressing SK-N-AS cells, neuronal differentiation is mainly observed in IMR-32 cells.
Discussion
In this study, we show that expression of the majority of class I HOX genes is associated with clinical phenotypes of neuroblastoma. HOX genes play essential roles in morphogenesis and cell differentiation during embryonic development.1 As neuroblastoma is an embryonal tumor capable of differentiating into ganglioneuroma or ganglioneuroblastoma, it has been suggested that HOX transcription factors may be involved in these processes.20, 27 Accordingly, it has been reported that morphological differentiation of neuroblastoma cells upon treatment with RA is accompanied by an increase of expression of a number of HOX genes.20, 27, 28 In line with these results, we show here that the expression of most HOX genes is significantly associated with prognostic markers and clinical outcome in primary neuroblastoma, suggesting that HOX genes are essentially involved in neuroblastoma pathogenesis. The potential relevance of these genes is further emphasized by the finding that a HOX gene expression-based classifier was able to predict accurately neuroblastoma patient outcome independently of age, stage and MYCN status. We observed that HOX genes of the A, B and C clusters were mainly upregulated in favorable neuroblastoma, while genes of the D cluster, particularly HOXD3, HOXD8, HOXD9 and HOXD10, were upregulated in unfavorable tumors. These findings are in contrast to a recent study of Mao et al.,21 who investigated the prognostic values of a panel of HOX genes in primary neuroblastoma, and did not observe any correlation of expression and clinical outcome for most of these. The difference between the two studies might be explained by the fact that Mao and co-workers21 had examined a limited number of samples comprising metastasized neuroblastoma without MYCN amplification only,29 while a large tumor cohort representing the entire spectrum of the disease has been analyzed in our study.
Among all HOX genes, HOXC9 expression was most prominently associated with favorable prognostic markers and with clinical outcome. The prognostic impact of HOXC9 is underlined by the fact that this gene was selected in all predictive models generated during the training phase of the process of HOX classifier generation, and that it was one of 144 genes of a prognostic classifier that we have reported previously.22 Using gene expression data from our group and others, HOXC9 expression has in addition been described to be associated with the clinical neuroblastoma phenotype in a recent study.21 Notably, we observed that HOXC9 expression levels were higher in neuroblastomas, which had spontaneously regressed than in age-related stage 4 tumors, suggesting that this transcription factor might be involved in the enigmatic molecular process of spontaneous regression.
Spatial and temporal colinearity is a hallmark of clustered HOX genes.30, 31 HOX genes that are positioned closer to the 5′ end of the cluster, such as HOXC9, are expressed later and more posterior during early development. In accordance with the embryonic expression pattern of HOX genes, we here show a significant correlation of increased HOXC9 expression levels of neuroblastomas from abdominal sites in comparison to tumors of the neck and chest. This finding contrasts with previous speculations,21 which suggested that HOXC9 expression might be higher in tumors of the neck and thorax because of their association with a better outcome24, 25 and a more differentiated histology.32
We next questioned which genetic or epigenetic factors might influence HOXC9 expression in primary neuroblastoma. A gain of chromosome 12, which comprises the HOXC9 locus, was associated with elevated HOXC9 transcript levels, suggesting that a gene–dosage effect might account for HOXC9 expression differences in neuroblastoma. These findings are in line with previous reports showing that numerical chromosomal aberrations, including chromosome 12 gain,33, 34 are associated with lower stages35 and better outcome in neuroblastoma.36 By contrast, we neither detected hypermethylation of the promoter region of HOXC9 nor inactivating mutations in the HOXC9 locus as a potential cause of diminished HOXC9 expression in unfavorable tumors. Alternatively, other mechanisms, such as regulatory RNAs37 or upstream transcriptional regulators,19 may control HOXC9 expression in neuroblastoma.
In line with data reported by Mao et al.,21 we observed reduced proliferation and cell cycle arrest in the G0/G1 phase upon HOXC9 re-expression. Moreover, we substantiate that Hox-C9 may induce neuronal differentiation in a fraction of neuroblastomas. In addition, we observed downregulation of REST after HOXC9 re-expression. REST is a well-known master negative regulator of neurogenesis,38 and has been reported to be degraded upon RA-induced neuroblastoma cell differentiation.39 Taking these findings together, it seems likely that Hox-C9 promotes a more differentiated neuroblastoma phenotype by the downregulation of REST.
The strong effect of Hox-C9 re-expression on neuroblastoma growth arrest observed by us and others21 suggested that other mechanisms than cell cycle arrest alone may contribute to this phenotype. Accordingly, we here provide substantial evidence that Hox-C9 promotes apoptosis in neuroblastoma in general, as indicated by a strong increase in the sub-G1 fraction and an increase in TUNEL-positive cells after Hox-C9 induction in all three cell lines. We demonstrate that HOXC9 expression activates the intrinsic pathway of apoptosis, probably by increasing the BAX:BCL-2 ratio, which in turn leads to the release of cytochrome c from the mitochondria into the cytosol and activates the intrinsic cascade of caspases. Delayed activation of the naturally occurring process of programmed cell death during development of the peripheral nervous system has been suggested to represent the molecular basis of spontaneous regression in neuroblastoma.11, 12 Taking the correlation of HOXC9 expression with favorable outcome and spontaneous regression in neuroblastoma, its developmental significance and its proapoptotic function in neuroblastoma cells into account, we hypothesize that Hox-C9 may contribute to the process of spontaneous regression in neuroblastoma.
In summary, we here demonstrate that distinct HOX gene expression patterns are associated with clinical phenotypes of neuroblastoma. We show that HOXC9 expression does not only correlate with favorable prognostic markers and beneficial clinical outcome but also with spontaneous regression in infant neuroblastoma. Re-expression of HOXC9 in neuroblastoma cell lines consistently leads to strong growth arrest and activates the intrinsic pathway of apoptosis. Taken together, we conclude from our data that HOX genes may contribute to neuroblastoma pathogenesis, and that Hox-C9 might be involved in the molecular process of spontaneous regression.
Materials and Methods
Patient characteristics
Tumors of 649 neuroblastoma patients were analyzed. Stages were classified according to the International Neuroblastoma Staging System:40 stage 1, n=153; stage 2, n=113; stage 3, n=91; stage 4, n=214; and stage 4S, n=78. Primary tumors were localized in the abdomen (n=135), at the neck/chest (n=69) or the adrenal glands (n=276). The primary tumor localization for the remaining 169 patients was unknown. The subgroup of patients showing regression of neuroblastoma consisted of 43 infants (localized, n=14; stage 4S, n=29), in which the tumor manifestations showed unambiguous regression without any cytotoxic treatment.41 The neuroblastoma subgroups used for methylation and aCGH analyses consisted of 46 and 209 tumors, respectively. Risk estimation of corresponding patients was performed according to the International Neuroblastoma Risk Group (INRG) classification system42 (methylation: high risk, n=21; intermediate and low risk, n=24; unclassified, n=1; aCGH: high risk, n=56; intermediate and low risk, n=153).
Gene expression analysis
Single-color gene expression profiles from 649 neuroblastoma tumors and 2 neuroblastoma cell lines were generated using 44K oligonucleotide microarrays as described previously.43 Total RNA of HOXC9- and GFP-expressing SK-N-AS and IMR-32 cells was isolated at 0, 6, 12, 24, 48 and 96 h using Trizol (Invitrogen, Karlsruhe, Germany). To determine global differences in the expression profiles of HOXC9- and GFP-induced SK-N-AS and IMR-32 cells, mean expression levels of each gene between HOXC9-induced and control cells were compared. Gene Ontology Tree Machine (GOTM)44 was used to identify functional categories associated with the condition of the respective cell line (Supplementary Materials and Methods). All raw and normalized microarray data are available through the Gene Expression Omnibus database (Accession: GSE45480).
HOX gene expression-based classification
To develop a HOX gene expression-based classifier (HOX classifier), the nearest shrunken centroids method (PAM)45 was applied on HOX gene expression data from 75 neuroblastoma patients with maximal divergent clinical outcome.22 Classification performance in the training set was evaluated by a 10 times repeated 5-fold cross-validation. Probes that were included in at least 65% of all cross-validation models were selected as classifier probes.22 Classification accuracy of the final HOX classifier comprising 33 HOX genes was assessed in an independent validation set of 574 neuroblastoma patients using the support vector machine (SVM)46 algorithm. Multivariate Cox proportional hazard models based on EFS and OS were used to analyze the prognostic impact of this classifier. The factors age (<18 months versus >18 months), tumor stage (stages 1–3 and 4S versus stage 4), MYCN (normal versus amplified) and HOX classifier were fitted into a stepwise-backward selection model.
Analysis of genetic aberrations and promoter methylation of HOXC9
Oligonucleotide aCGH profiles from 209 neuroblastoma tumors were generated using 44K, 105K or 1M microarrays as described previously.35, 47 Array-CGH analysis was preformed as described previously47 and visualized using the Integrative Genomics Viewer.48 Because of technical reasons, 9 out of 209 aCGH profiles could not be visualized. Array-CGH data are available at Gene Expression Omnibus (Accession: GSE25771 and GSE45480). HOXC9 promoter methylation analysis was performed by Sequenom Inc. (Hamburg, Germany; Supplementary Materials and Methods and Supplementary Table S5). Whole genome-amplified DNA from primary neuroblastoma specimens and neuroblastoma cell lines were used to sequence amplicons covering the first 500 nucleotides upstream of the HOXC9 start site, 5′- and 3′-UTRs and the complete coding region with exon–intron boundaries. Sequencing was performed by Beckman Coulter Genomics (Danvers, MA, USA; Supplementary Materials and Methods and Supplementary Table S5).
Data analysis and statistics
Statistical analysis of associations between HOX mRNA expression levels and neuroblastoma subgroups, prognostic markers and outcome in neuroblastoma was performed using SPSS version 20.0 (IBM, Mainz, Germany). Two-tailed non-parametric tests (Mann–Whitney U-test and Kruskal–Wallis test) were used where appropriate. All HOX genes were first tested in a univariable Cox regression model based on OS and EFS. For all HOX genes with P-values ≤0.05, the whole cohort (n=649) was divided using quartiles based on HOX gene expression levels. Kaplan–Meier estimates for OS (n=649) and EFS (n=628) were compared by log-rank test. In addition, HOXC9 was investigated separately by using quartile values of HOXC9 expression levels in a set of 244 neuroblastoma tumors. These values were applied on an independent set of 405 neuroblastoma expression profiles as a cutoff for high, intermediate–high, intermediate–low and low HOXC9 expressions. Kaplan–Meier estimates for OS (n=405) and EFS (n=384) were compared by log-rank test. Recurrence, progression and death from disease were considered as events. Multivariate Cox proportional hazard models based on EFS and OS were used to analyze the prognostic impact of HOXC9 expression in neuroblastoma. The factors age (>18 months versus <18 months), tumor stage (stage 4 versus stages 1–3 and 4S), MYCN (amplified versus normal) and HOXC9 expression were fitted into a stepwise-backward selection model. The likelihood ratio test P for inclusion was ≤0.05 and for exclusion was >0.05. Quantitative data for functional analysis were shown as means±S.D. Unpaired two-tailed Student’s t-tests were used where appropriate.
Cell culture
SK-N-AS and CHP-212 cells were obtained from ATCC (Manassas, VA, USA) and IMR-32 cells were purchased from German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). Neuroblastoma cell lines were authenticated at the DSMZ. Cells were maintained as described in Supplementary Materials and Methods.
Retroviral plasmids and stable inducible neuroblastoma cell lines
A human HOXC9 full open reading frame was obtained from the retroviral vector pBIG2rHOXC9 (kind gift of Malte Buchholz, Philipps-University of Marburg, Marburg, Germany) and cloned into the pRevTRE vector (Clontech, Heidelberg, Germany; Supplementary Materials and Methods). Polyclonal neuroblastoma cell lines stably expressing either HOXC9 or GFP under the control of the reverse tetracycline-controlled transactivator were generated using the RevTet System (Clontech) according to the manufacturer’s instructions with marginal modifications (Supplementary Materials and Methods).
Expression analyses using immunoblots and qRT-PCR
Immunoblots were prepared as described previously.49 Cytosolic cell extracts were isolated as described elsewhere.50 Total protein extracts were isolated either with RIPA or Cell Lysis Buffer (Biovision, Hannover, Germany). Densitometric quantification of immunoblots was performed using the Image J software (NIH, Bethesda, MD, USA).51 qRT-PCR was carried out two times in duplicates as described previously.52 Antibody and primer information is given in Supplementary Materials and Methods and Supplementary Table S5, respectively.
In vitro growth assays
Cell viability was analyzed by Trypan blue dye exclusion (Supplementary Materials and Methods) and cell cycle distribution was assessed by FACS analysis as described previously.53
Tumorigenicity assays
Soft agar assays were used to assess the effect of HOXC9 on colony formation ability of neuroblastoma cells (Supplementary Materials and Methods). A total of 32 six-week-old female athymic nude-Foxn1nu mice (Harlan Laboratories, Horst, The Netherlands) were used for the establishment of neuroblastoma xenograft tumors (Supplementary Materials and Methods).
Cell viability analysis
The TUNEL analysis was performed using the In Situ Cell Death Detection Kit according to the manufacturer’s protocol (Roche, Mannheim, Germany; Supplementary Materials and Methods). Annexin-V-binding analysis was carried out using APC-coupled Annexin-V and 7-AAD according to the manufacturer’s protocol (BD Biosciences, Heidelberg, Germany). Apoptotic cells were detected by FACS (FACS Canto; BD Biosciences) using the DIVA software (BD Biosciences). The extent of DNA fragmentation was evaluated in a hypoploidy assay as described elsewhere.54
Abbreviations
- ABD:
-
abdomen
- Aberr:
-
aberration
- aCGH:
-
array-comparative genomic hybridization
- AG:
-
adrenal glands
- amp:
-
amplification
- C:
-
chest
- Dox:
-
doxycycline
- DSMZ:
-
German Collection of Microorganisms and Cell Cultures
- EFS:
-
event-free survival
- fav:
-
favorable
- GO:
-
gene ontology
- GOTM:
-
Gene Ontology Tree Machine
- Hox:
-
homeobox
- IGV:
-
integrative genomics viewer
- Loc:
-
localization
- NK:
-
neck
- norm:
-
normal
- OS:
-
overall survival
- PAM:
-
prediction analysis for microarrays
- RA:
-
retinoic acid
- SR:
-
spontaneously regressive
- SVM:
-
support vector machine
- unfav:
-
unfavorable
References
Gehring WJ, Hiromi Y . Homeotic genes and the homeobox. Annu Rev Genet 1986; 20: 147–173.
Bel-Vialar S, Itasaki N, Krumlauf R . Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development 2002; 129: 5103–5115.
Nordstrom U, Maier E, Jessell TM, Edlund T . An early role for WNT signaling in specifying neural patterns of Cdx and Hox gene expression and motor neuron subtype identity. PLoS Biol 2006; 4: e252.
Daniels TR, Neacato II, Rodriguez JA, Pandha HS, Morgan R, Penichet ML . Disruption of HOX activity leads to cell death that can be enhanced by the interference of iron uptake in malignant B cells. Leukemia 2010; 24: 1555–1565.
Shears L, Plowright L, Harrington K, Pandha HS, Morgan R . Disrupting the interaction between HOX and PBX causes necrotic and apoptotic cell death in the renal cancer lines CaKi-2 and 769-P. J Urol 2008; 180: 2196–2201.
Plowright L, Harrington KJ, Pandha HS, Morgan R . HOX transcription factors are potential therapeutic targets in non-small-cell lung cancer (targeting HOX genes in lung cancer). Br J Cancer 2009; 100: 470–475.
Abate-Shen C . Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer 2002; 2: 777–785.
Shimada H, Ambros IM, Dehner LP, Hata J, Joshi VV, Roald B . Terminology and morphologic criteria of neuroblastic tumors: recommendations by the International Neuroblastoma Pathology Committee. Cancer 1999; 86: 349–363.
Nakagawara A, Arima-Nakagawara M, Scavarda NJ, Azar CG, Cantor AB, Brodeur GM . Association between high levels of expression of the TRK gene and favorable outcome in human neuroblastoma. N Engl J Med 1993; 328: 847–854.
van Noesel MM . Neuroblastoma stage 4S: a multifocal stem-cell disease of the developing neural crest. Lancet Oncol Mar 2012; 13: 229–230.
Pritchard J, Hickman JA . Why does stage 4s neuroblastoma regress spontaneously? Lancet 1994; 344: 869–870.
Nakagawara A . Neural crest development and neuroblastoma: the genetic and biological link. Prog Brain Res 2004; 146: 233–242.
Nakagawara A, Ohira M . Comprehensive genomics linking between neural development and cancer: neuroblastoma as a model. Cancer Lett 2004; 204: 213–224.
Fischer M, Oberthuer A, Brors B, Kahlert Y, Skowron M, Voth H et al. Differential expression of neuronal genes defines subtypes of disseminated neuroblastoma with favorable and unfavorable outcome. Clin Cancer Res 2006; 12: 5118–5128.
Mosse YP, Laudenslager M, Khazi D, Carlisle AJ, Winter CL, Rappaport E et al. Germline PHOX2B mutation in hereditary neuroblastoma. Am J Hum Genet 2004; 75: 727–730.
Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF . The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature 1999; 399: 366–370.
Schwab M, Alitalo K, Klempnauer KH, Varmus HE, Bishop JM, Gilbert F et al. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature 1983; 305: 245–248.
Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, Wong KY et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med 1985; 313: 1111–1116.
Alaminos M, Mora J, Cheung NK, Smith A, Qin J, Chen L et al. Genome-wide analysis of gene expression associated with MYCN in human neuroblastoma. Cancer Res 2003; 63: 4538–4546.
Manohar CF, Furtado MR, Salwen HR, Cohn SL . Hox gene expression in differentiating human neuroblastoma cells. Biochem Mol Biol Int 1993; 30: 733–741.
Mao L, Ding J, Zha Y, Yang L, McCarthy BA, King W et al. HOXC9 links cell-cycle exit and neuronal differentiation and is a prognostic marker in neuroblastoma. Cancer Res 2011; 71: 4314–4324.
Oberthuer A, Berthold F, Warnat P, Hero B, Kahlert Y, Spitz R et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol 2006; 24: 5070–5078.
Oberthuer A, Hero B, Berthold F, Juraeva D, Faldum A, Kahlert Y et al. Prognostic impact of gene expression-based classification for neuroblastoma. J Clin Oncol 2010; 28: 3506–3515.
Hughes M, Marsden HB, Palmer MK . Histologic patterns of neuroblastoma related to prognosis and clinical staging. Cancer 1974; 34: 1706–1711.
Shimada H, Chatten J, Newton WA Jr., Sachs N, Hamoudi AB, Chiba T et al. Histopathologic prognostic factors in neuroblastic tumors: definition of subtypes of ganglioneuroblastoma and an age-linked classification of neuroblastomas. J Natl Cancer Inst 1984; 73: 405–416.
Liu X, Kim CN, Yang J, Jemmerson R, Wang X . Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 1996; 86: 147–157.
Peverali FA, D’Esposito M, Acampora D, Bunone G, Negri M, Faiella A et al. Expression of HOX homeogenes in human neuroblastoma cell culture lines. Differentiation 1990; 45: 61–69.
Merrill RA, Ahrens JM, Kaiser ME, Federhart KS, Poon VY, Clagett-Dame M . All-trans retinoic acid-responsive genes identified in the human SH-SY5Y neuroblastoma cell line and their regulated expression in the nervous system of early embryos. Biol Chem 2004; 385: 605–614.
Asgharzadeh S, Pique-Regi R, Sposto R, Wang H, Yang Y, Shimada H et al. Prognostic significance of gene expression profiles of metastatic neuroblastomas lacking MYCN gene amplification. J Natl Cancer Inst 2006; 98: 1193–1203.
Graham A, Papalopulu N, Krumlauf R . The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell 1989; 57: 367–378.
Lewis EB . A gene complex controlling segmentation in Drosophila. Nature 1978; 276: 565–570.
Joshi VV, Cantor AB, Altshuler G, Larkin EW, Neill JS, Shuster JJ et al. Recommendations for modification of terminology of neuroblastic tumors and prognostic significance of Shimada classification. A clinicopathologic study of 213 cases from the Pediatric Oncology Group. Cancer 1992; 69: 2183–2196.
Chen QR, Bilke S, Wei JS, Whiteford CC, Cenacchi N, Krasnoselsky AL et al. cDNA array-CGH profiling identifies genomic alterations specific to stage and MYCN-amplification in neuroblastoma. BMC Genom 2004; 5: 70.
Coco S, Theissen J, Scaruffi P, Stigliani S, Moretti S, Oberthuer A et al. Age-dependent accumulation of genomic aberrations and deregulation of cell cycle and telomerase genes in metastatic neuroblastoma. Int J Cancer 2012; 131: 1591–1600.
Spitz R, Oberthuer A, Zapatka M, Brors B, Hero B, Ernestus K et al. Oligonucleotide array-based comparative genomic hybridization (aCGH) of 90 neuroblastomas reveals aberration patterns closely associated with relapse pattern and outcome. Genes Chromosomes Cancer 2006; 45: 1130–1142.
Janoueix-Lerosey I, Schleiermacher G, Michels E, Mosseri V, Ribeiro A, Lequin D et al. Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol 2009; 27: 1026–1033.
Yekta S, Tabin CJ, Bartel DP . MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat Rev Genet 2008; 9: 789–796.
Gao Z, Ure K, Ding P, Nashaat M, Yuan L, Ma J et al. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J Neurosci 2011; 31: 9772–9786.
Singh A, Rokes C, Gireud M, Fletcher S, Baumgartner J, Fuller G et al. Retinoic acid induces REST degradation and neuronal differentiation by modulating the expression of SCF(beta-TRCP) in neuroblastoma cells. Cancer 2011; 117: 5189–5202.
Brodeur GM, Pritchard J, Berthold F, Carlsen NL, Castel V, Castelberry RP et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J Clin Oncol 1993; 11: 1466–1477.
Hero B, Simon T, Spitz R, Ernestus K, Gnekow AK, Scheel-Walter HG et al. Localized infant neuroblastomas often show spontaneous regression: results of the prospective trials NB95-S and NB97. J Clin Oncol 2008; 26: 1504–1510.
Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol 2009; 27: 289–297.
Oberthuer A, Juraeva D, Li L, Kahlert Y, Westermann F, Eils R et al. Comparison of performance of one-color and two-color gene-expression analyses in predicting clinical endpoints of neuroblastoma patients. Pharmacogenomics J 2010; 10: 258–266.
Zhang B, Schmoyer D, Kirov S, Snoddy J . GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinform 2004; 5: 16.
Tibshirani R, Hastie T, Narasimhan B, Chu G . Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA 2002; 99: 6567–6572.
Vapnik VN . An overview of statistical learning theory. IEEE Trans Neural Netw 1999; 10: 988–999.
Fischer M, Bauer T, Oberthur A, Hero B, Theissen J, Ehrich M et al. Integrated genomic profiling identifies two distinct molecular subtypes with divergent outcome in neuroblastoma with loss of chromosome 11q. Oncogene 2009; 29: 865–875.
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G et al. Integrative genomics viewer. Nat Biotechnol 2011; 29: 24–26.
Ackermann S, Goeser F, Schulte JH, Schramm A, Ehemann V, Hero B et al. Polo-like kinase 1 is a therapeutic target in high-risk neuroblastoma. Clin Cancer Res 2011; 17: 731–741.
Kashkar H, Kronke M, Jurgensmeier JM . Defective Bax activation in Hodgkin B-cell lines confers resistance to staurosporine-induced apoptosis. Cell Death Differ 2002; 9: 750–757.
Schneider CA, Rasband WS, Eliceiri KW . NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9: 671–675.
Fischer M, Skowron M, Berthold F . Reliable transcript quantification by real-time reverse transcriptase-polymerase chain reaction in primary neuroblastoma using normalization to averaged expression levels of the control genes HPRT1 and SDHA. J Mol Diagn 2005; 7: 89–96.
Ehemann V, Hashemi B, Lange A, Otto HF . Flow cytometric DNA analysis and chromosomal aberrations in malignant glioblastomas. Cancer Lett 1999; 138: 101–106.
Nicoletti I, Migliorati G, Pagliacci MC, Grignani F, Riccardi C . A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Methods 1991; 139: 271–279.
Acknowledgements
We thank Anne Engesser and Hafiza Fakhera Sherkheli for excellent technical assistance, Falk Hertwig for critical reading of the manuscript, Ruth Volland for statistical support and Jens Seeger for fruitful discussions. This work was supported by grants from the Cologne Center for Molecular Medicine, the Bundesministerium für Bildung und Forschung (BMBF) through the National Genome Research Network plus (NGFNplus, Grant No. 01GS0895), the Fördergesellschaft Kinderkrebs-Neuroblastom-Forschung e.V. and by the Competence Network Pediatric Oncology and Hematology (KPOH).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Raschellá
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Kocak, H., Ackermann, S., Hero, B. et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis 4, e586 (2013). https://doi.org/10.1038/cddis.2013.84
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.84
Keywords
This article is cited by
-
MYCN and SNRPD3 cooperate to maintain a balance of alternative splicing events that drives neuroblastoma progression
Oncogene (2024)
-
Development and validation of a 21-gene prognostic signature in neuroblastoma
Scientific Reports (2023)
-
Identification of immune-related signature with prognosis in children with stage 4 and 4S neuroblastoma
Clinical and Translational Oncology (2023)
-
SOX11 regulates SWI/SNF complex components as member of the adrenergic neuroblastoma core regulatory circuitry
Nature Communications (2023)
-
Expression and activation of nuclear hormone receptors result in neuronal differentiation and favorable prognosis in neuroblastoma
Journal of Experimental & Clinical Cancer Research (2022)