Abstract
Jasmonic acid (JA) is an important phytohormone that regulates plant defense responses against herbivore attack, pathogen infection and mechanical wounding. In this report, we provided biochemical and genetic evidence to show that the Arabidopsis thaliana NAC family proteins ANAC019 and ANAC055 might function as transcription activators to regulate JA-induced expression of defense genes. The role of the two NAC genes in JA signaling was examined with the anac019 anac055 double mutant and with transgenic plants overexpressing ANAC019 or ANAC055. The anac019 anac055 double mutant plants showed attenuated JA-induced VEGETATIVE STORAGE PROTEIN1 (VSP1) and LIPOXYGENASE2 (LOX2) expression, whereas transgenic plants overexpressing the two NAC genes showed enhanced JA-induced VSP1 and LOX2 expression. That the JA-induced expression of the two NAC genes depends on the function of COI1 and AtMYC2, together with the finding that overexpression of ANAC019 partially rescued the JA-related phenotype of the atmyc2-2 mutant, has led us to a hypothesis that the two NAC proteins act downstream of AtMYC2 to regulate JA-signaled defense responses. Further evidence to substantiate this idea comes from the observation that the response of the anac019 anac055 double mutant to a necrotrophic fungus showed high similarity to that of the atmyc2-2 mutant.
Similar content being viewed by others
Introduction
The jasmonate family of oxylipins, including jasmonic acid (JA), methyl jasmonic acid (MeJA) and other bioactive derivatives of JA are important signaling molecules in the plant kingdom. JAs are perhaps best known for their role in regulating defense responses against biotic stresses such as herbivore attack and necrotrophic pathogen infection 1, 2, 3, 4, 5, 6. In addition to biotic stresses, JAs are also involved in the control of plant responses to a range of abiotic stresses 7, 8, 9, 10, 11, 12.
Molecular genetic studies, mainly conducted in the Arabidopsis thaliana model system, have identified several important players in the JA signal transduction pathway. The coronatine insensitive 1 (coi1) mutant in Arabidopsis is fully insensitive to JA in both root growth inhibition and defense gene expression 13. Molecular characterization of COI1 has indicated that this gene encodes an F-box protein, which suggests the involvement of a ubiquitin-mediated protein-degradation process in JA signaling 14. Support for this hypothesis comes from the demonstration that COI1 interacts with Skp1 and Cullin1 to assemble a functional SCFCOI1 ubiquitin ligase complex in vivo 15, 16. Compared with coi1, the jasmonate resistance1 (jar1) 17 and jasmonate insensitive1 (jin1) 18, 19 mutants exhibit a relatively weak phenotype in JA-induced inhibition of root growth. JAR1 encodes an enzyme that has JA adenylation activity to form JA-amino acid conjugates, especially JA-isoleucine (JA-Ile), which suggests that JA-Ile, rather than JA itself, might be the active hormone 20. JIN1 encodes a nuclear-localized basic helix-loop-helix (bHLH)-type transcription factor known as AtMYC2 19. Genome-wide transcriptional profiling demonstrated that AtMYC2 acts as both activator and repressor to regulate diverse aspects of JA responses 21. Recently, a family of Arabidopsis proteins named JAZ (jasmonate ZIM domain) was identified as a target of the SCFCOI1 ubiquitin ligase complex in JA signaling 22, 23. Significantly, JA-Ile promotes the binding of the SCFCOI1 ubiquitin ligase to and subsequent degradation of the JAZ proteins, which suggests that the SCFCOI1-JAZ complex may be the site of JA-Ile perception 23. Furthermore, JAZ proteins interact and negatively regulate AtMYC2, a central regulator of JA-induced gene expression. Therefore, the JAZ family proteins represent the molecular link between SCFCOI1-mediated protein degradation and transcriptional activation of jasmonate responses 22.
Among the identified components of JA signaling, the role of JIN1/AtMYC2 (henceforth referred to as AtMYC2) is interesting. As mentioned above, AtMYC2 differentially regulates two types of JA signaling defense responses 19, 24, 25. One of these types, which is positively regulated by AtMYC2, induces the expression of genes that are involved in the response to wounding (mechanical or biotic). The other type, which is negatively regulated by AtMYC2, represses the expression of pathogen defense-related genes. Therefore, while the jin1/atmyc2 mutants showed decreased expression of wound responsive genes such as VEGETATIVE STORAGE PROTEIN1 (VSP1) 18 and LIPOXYGENASE2 (LOX2) 26, they showed increased expression of a group of pathogen defensive genes that includes PDF1.2 27 and PATHOGENESIS RELATED1 (PR-1) 19, 24, 25. However, less is known about the JA signaling components that function downstream of AtMYC2.
NAC (NAM/ATAF1, 2/CUC2) proteins are a family of plant-specific transcription factors with diverse biological functions 28. Accumulating evidence indicates that NAC family proteins play an important role in different aspects of plant development 29, 30, 31, 32. In addition, several NAC proteins in Arabidopsis are believed to be important in plant responses to abiotic stresses. For example, it was shown that three dehydration-inducible NAC genes (ANAC019, ANAC055 and ANAC072) were associated with drought tolerance 33 and that AtNAC2/ANAC092 was associated with salt-stress responses 34. A role for NAC proteins in biotic stress responses was recently exemplified by the demonstration that the Arabidopsis NAC protein ATAF2 29 functions as a repressor of pathogenesis-related genes 35. However, the biological functions for most of the members of the NAC-family transcription factors remain to be determined 28.
ANAC019 and ANAC055 are closely related NAC family proteins in Arabidopsis 33, 36. In this report, we provided evidence to show that ANAC019 and ANAC055 may function as transcription activators to regulate JA-induced expression of defense genes. The role of the two NAC genes in JA signaling was examined with the anac019 anac055 double mutant and transgenic plants overexpressing ANAC019 or ANAC055. Our data support a hypothesis that the two NAC proteins act downstream of AtMYC2 to regulate JA-signaled defense responses.
Results
ANAC019 and ANAC055 are JA-inducible genes encoding NAC family proteins
We identified At1g52890 and At3g15500 as JA-inducible genes in our microarray analyses using the Arabidopsis whole genome chip (Affymetrix) 37. Both At1g52890 and At3g15500 encode proteins of 317 amino acids and their N-terminal regions contain an NAC domain that is highly similar to all members of the plant-specific NAC family proteins 36. At1g52890 and At3g15500 were therefore re-named ANAC019 and ANAC055, respectively, according to the nomenclature that has been established for the family of Arabidopsis NAC proteins 36. At the amino-acid level, ANAC019 showed 72% sequence similarity to ANAC055 and the regions of homology were observed not only in the NAC domain but also in the C-terminus 33, which suggests that the two members may have similar or overlapping biological functions.
ANAC019 and ANAC055 expression is induced by MeJA in a COI1- and AtMYC2-dependent manner
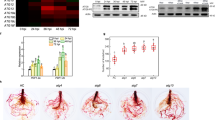
The MeJA-induced expression of ANAC019 and ANAC055 was verified using RNA gel blot analysis. As shown in Figure 1A, transcripts of the two NAC genes were undetectable in the absence of MeJA but were induced upon MeJA treatment. An increase in the number of ANAC019 and ANAC055 transcripts was detected as early as 15 min after applying MeJA, and this increase in transcript number was maintained up to 6 h after treatment.
MeJA-induced expression of ANAC019 and ANAC055. (A) MeJA-induced expression of the two NAC genes in wild-type plants. (B) MeJA-induced expression of the two NAC genes in different mutant backgrounds. Two-week-old plants were treated with 50 μM MeJA and tissues were collected at the indicated times for RNA extraction. In all, 30 μg of total RNA was loaded per lane, and the blot was hybridized with the indicated probes. A duplicated gel stained with EtBr was used as a loading control.
The MeJA-induced expression of the two NAC genes was compared among the wild-type and two JA-related mutants, coi1-1 14 and atmyc2-2 25. As shown in Figure 1B, the MeJA-induced expression levels of ANAC019 and ANAC055 were significantly reduced in coi1-1 and atmyc2-2. These results show that the MeJA-induced activation of ANAC019 and ANAC055 expression requires the function of the COI1 and AtMYC2 proteins, two of the known essential components of JA signaling in Arabidopsis.
ANAC019 transcription factor binds directly to the VSP1 promoter
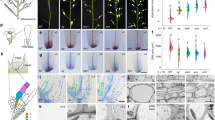
Tran et al. 33 demonstrated that ANAC019 and ANAC055 act as transcription factors to activate the expression of drought-inducible genes. Furthermore, they determined the complete DNA-binding sequence that is recognized by the two NAC proteins, which include the so-called NAC recognition sequence (NACRS) CATGT and the core-binding site CACG. As shown in Supplementary information, Figure S1, our sequence examination revealed two CATGT motifs and six CACG motifs in the promoter region of VSP1, a well-recognized marker gene for JA-induced responses in Arabidopsis 18. To verify the possibility that ANAC019 might bind the promoter of VSP1, we tested the binding activity of ANAC019 to a 10-bp DNA segment (CATGTCCACG) at positions −369 to −360 relative to the predicted translation start codon of VSP1 38. The 10-bp cis-element CATGTCCACG contains a NACRS and a core-binding site, which are organized as a tandem array spaced by a C (Figure 2A and Supplementary information, Figure S1). Purified His-ANAC019 fusion protein was incubated with a labeled DNA probe containing four tandem repeats of the 10-bp cis-element (named as vsp, Figure 2A) and incubation mixtures were then analyzed using a gel mobility shift assay. A high-affinity DNA-protein complex was detected along with the free probe (Figure 2B, lane 2), and the DNA-binding activity was reduced by the addition of an excess of unlabeled vsp oligonucleotide probe (Figure 2B, lanes 6-8). To define the sequence specificity of the DNA-binding activities, we analyzed the binding activities of the ANAC019 fusion protein to three mutant versions of the vsp probe (named as m1, m2 and m3, respectively, Figure 2A). When DNA probes with mutations in only CATGT (substituted by AAAAA) or CACG (substituted by AAAA) were tested, no apparent reduction in binding activity was observed (Figure 2B, lanes 3 and 4). By contrast, when both CATGT and CACG were mutated, ANAC019 lost its binding activity (Figure 2B, lane 5). Furthermore, unlabeled mutant probe in which both CATGT and CACG were substituted lost its ability to compete with the wild-type vsp probe for interaction with the ANAC019 fusion protein (Figure 2B, lane 9). These results suggest that ANAC019 can interact directly with the promoter of VSP1. Given the high sequence similarity of ANAC055 with that of ANAC019, it is reasonable to predict that ANAC055 also can bind directly to the VSP1 promoter.
In vitro binding of His-ANAC019 to the CATGT and CACG motifs in the promoter of the Arabidopsis VSP1 gene. (A) Oligonucleotides used in the gel mobility shift assay. The vsp probe contains four tandem repeats of a cis-element containing a NAC recognition site CATGT (underlined) and a core-binding site CACG (double underlined), which present in the promoter region of VSP1. To demonstrate the specificity of ANAC019 binding to the VSP1 promoter, three mutant versions of the vsp probe were used: m1, CATGT was substituted by AAAAA; m2, CACG was substituted by AAAA; m3, both CATGT and CACG were mutated. (B) A gel mobility shift assay to test the binding of His-ANAC019 to the CATGT and CACG motifs in the promoter of the VSP1 gene. Protein-DNA complexes were detected when His-ANAC019 fusions were incubated with vsp (lane 2), m1 (lane 3), m2 (lane 4), but not with m3 (lane 5). m3 lost its ability to compete with the binding of His-ANAC019 to vsp (lane 9). Binding reactions contained approximately 300 ng of His-ANAC019 fusion protein and 0.05 pmol/μL of labeled probe. No protein was added in lane 1. The triangle indicates the addition of 10× (lane 6), 50× (lane 7) and 100× (lane 8) molar excess amounts of unlabeled vsp competitors (Comp.). The arrow indicates the protein-DNA complex. FP, free probe.
Nuclear localization of ANAC019 and ANAC055
To investigate the subcellular localization of the two NAC proteins, we generated transgenic plants that contained full-length cDNA of ANAC019 or ANAC055 fused with the cDNA of the green fluorescent protein (GFP) under the control of the 35S promoter. Homozygous 35S:ANAC019-GFP and 35S:ANAC055-GFP plants showed increased MeJA-induced VSP1 expression compared to 35S:GFP plants (data not shown), which suggests that the ANAC019-GFP and ANAC055-GFP fusions are functional. Root tips of the transgenic seedlings were examined for localization of GFP fluorescence. Root tips of transgenic plants containing the 35S:GFP vector control were found to display a broad distribution of GFP fluorescence throughout the cells (Figure 3, upper panel). By contrast, the fusion proteins ANAC019-GFP (Figure 3, middle panel) and ANAC055-GFP (Figure 3, bottom panel) were found to target the nuclei. These results suggest that ANAC019 and ANAC055 can be localized to the nucleus.
Subcellular localization of the NAC-GFP fusion proteins. Root tips of transgenic plants containing the 35S:GFP (upper panel), 35S:ANAC019-GFP (middle panel) and 35S:ANAC055-GFP (bottom panel) fusion genes were observed and photographed using laser-scanning confocal microscope (LSM 510, Zeiss, Oberkochen, Germany).
ANAC019 and ANAC055 C-terminal domains have transactivational activity
Until now it was generally believed that the C-terminal domain of NAC family proteins had transactivational activity 32, 33, 34, 35, 36. Here, we tested for the presence of transactivation domains in ANAC019 and ANAC055 using yeast as an assay system. Yeast assays were conducted using the previously described yeast strain HF7c, which contains the two reporter genes His3 and LacZ 32. The C-terminal domains (amino acids 148-317) of ANAC019 and ANAC055, respectively, were fused to the GAL4 DNA-binding domain to examine their ability to activate transcription from the GAL4 upstream activation sequence (UAS) and thereby promote yeast growth. The yeast cells containing pBD-ANAC019C, pBD-ANAC055C and control plasmid pBD all grew well on YPDA medium (Figure 4B), while on SD medium without histidine and tryptophan only yeast cells containing pBD-ANAC019C or pBD-ANAC055C could grow (Figure 4C), which indicates that ANAC019C and ANAC055C have transcription activation activity and therefore could promote yeast growth without histidine and trytophan. In the β-gal activity assay, the yeast cells containing pBD-ANAC019C or pBD-ANAC055C also turned blue, indicating the activation of another reporter gene LacZ (Figure 4D). These results indicate that the C-terminal domains of ANAC019 and ANAC055 have transactivational activity in yeast.
Yeast assays showing the transcriptional activity and dimerization of ANAC019 and ANAC055. (A-D) The C-terminal domains of ANAC019 and ANAC055 showed transcription activation activity. Yeast cells were transformed with the indicated plasmids (A). The transformants were streaked on YPDA (B) and SD/Trp−His− medium to examine growth (C) and assayed for β-galactosidase activity on replica filters (D). (E-H) Dimerization of ANAC019 and ANAC055 analyzed using yeast two-hybrid assays. Yeast cells were co-transformed with the indicated plasmids (E). The transformants were streaked on YPDA (F) and SD/Trp−Leu−His− medium to examine growth (G) and assayed for β-galactosidase activity on replica filters (H).
Dimerization of ANAC019 and ANAC055
To test the ability of ANAC019 and ANAC055 to dimerize using a yeast two-hybrid assay, yeast cells were co-transformed with different combinations of constructs (Figure 4E). The results indicate that, whereas all the yeast lines grew well on YPDA medium (Figure 4F), only the yeast lines containing the construct combinations of pAD-ANAC019+pBD-ANAC019 and pAD-ANAC055+pBD-ANAC055 could grow well on SD/His− medium plus 10 mM amino-1, 2, 4-triazole (3-AT) and turned blue during a β-gal activity assay (Figure 4G–4H). This suggests that ANAC019 or ANAC055 interacts with itself in yeast. However, and on the contrary, yeast lines containing the construct combinations of pAD-ANAC019+pBD-ANAC055 and pAD-ANAC055+pBD-ANAC019 could grow fewer clones on the SD/His− medium plus 10 mM 3-AT (Figure 4G), and these clones failed to show β-gal activity (Figure 4H). This suggests that ANAC019 and ANAC055 cannot interact in yeast. As a control, yeast lines containing the pAD+pBD-ANAC019 or the pAD+pBD-ANAC055 construct could not grow (Figure 4G–4H). Collectively, the results from the yeast assays suggest that, for ANAC019 and ANAC055, homodimeric complexes are formed preferentially over heterodimeric complexes. This dimerization assay may help to explain why the anac019 anac055 double mutant shows stronger JA-related phenotypes than the single mutants (see below).
The anac019 anac055 double mutant shows reduced expression of MeJA-induced VSP1 and LOX2
In order to explore the physiological functions of the two NAC genes in JA signaling, we identified and analyzed the Arabidopsis T-DNA insertion lines anac019 and anac055 (see Materials and Methods). The anac019 allele (SALK_096295) contains a T-DNA insertion in the third exon of ANAC019 (Figure 5A). The anac055 allele (SALK_014331) contains a T-DNA insertion in the third exon of ANAC055 (Figure 5A). However, compared with the wild type, neither anac019 nor anac055 showed altered JA-induced responses in either root growth or defense gene expression (data not shown). Therefore, anac019 and anac055 were crossed and an F2 population that was segregated according to the two T-DNA insertions was obtained. Through PCR-based genotyping of 266 individuals from this F2 population, we obtained 15 (close to 1/16) anac019 anac055 double mutant plants. These results demonstrate that each of the T-DNAs had segregated as a single locus, and that both anac019 and anac055 are single-locus T-DNA insertion mutants. RNA gel blot analysis indicated that the double mutants simultaneously disrupted ANAC019 and ANAC055 expression (Figure 5B).
Reduced expression of VSP1 and LOX2 in response to MeJA in the anac019 anac055 double mutant. (A) Diagrams of ANAC019 and ANAC055 showing the positions of the T-DNA insertions. (B) RNA gel blot analysis showing disrupted expression of the NAC genes in the anac019 anac055 double mutant. Two-week-old plants were treated with 50 μM MeJA and tissues were collected at the indicated times for RNA extraction. In all, 30 μg of total RNA was loaded per lane. A duplicated gel stained with EtBr was used as a loading control. (C) Comparison of seedling root length of the indicated genotypes in different concentrations of MeJA. Each data point represents the mean ± SD of 20 plants per genotype. Three independent experiments were conducted and similar results were obtained. (D) Comparison of MeJA-induced expression of VSP1 and LOX2 in the anac019 anac055 double mutant and the wild type. Two-week-old seedlings were treated with 50 μM MeJA and tissues were collected at the indicated times for RNA extraction. In all, 10 μg of total RNA was loaded per lane. A duplicated gel stained with EtBr was used as a loading control.
The MeJA-induced response of the anac019 anac055 double mutant was first examined using a root growth inhibition assay. As shown in Figure 5C, in the absence or presence of a range of concentrations of MeJA, the root length of the anac019 anac055 double mutant was comparable with that of the wild type. To further study the effects of the anac019 anac055 double mutant on JA-regulated gene expression, we analyzed the expression levels of VSP1 and LOX2, which are two marker genes for JA-regulated defense responses in Arabidopsis. RNA gel blot analyses indicated that, in the absence of MeJA induction, the transcripts of the two genes were barely detectable (Figure 5D). MeJA treatment induced the expression of the two genes in both the wild-type and the anac019 anac055 double mutant, and the transcript accumulation levels of VSP1 and LOX2 were found to be lower in the double mutant than in the wild type (Figure 5D). These results indicate that the disruption of ANAC019 and ANAC055 expression leads to the attenuation of MeJA-induced VSP1 and LOX2 expression and suggest that there is a significant functional redundancy between ANAC019 and ANAC055.
Overexpression of ANAC019 and ANAC055 enhances MeJA-induced expression of VSP1 and LOX2
The effects of the NAC genes on JA signaling were also examined in transgenic Arabidopsis plants that overexpressed the full-length cDNAs of ANAC019 or ANAC055 under the control of the 35S promoter (see Materials and Methods). Homozygous transgenic lines were examined by RNA gel blot analysis for elevated expression of the ANAC019 (Figure 6A) or ANAC055 (Figure 6C) transcripts. Representative lines named 35S:ANAC019-1# and 35S:ANAC055-4#, respectively, were selected to compare their responses to MeJA with those of the wild type.
Increased expression of VSP1 and LOX2 in response to MeJA, due to overexpression of the two NAC genes. (A) RNA gel blot analysis showing enhanced expression of ANAC019 in different ANAC019-overexpressing lines. Tissues of 2-week-old seedlings (untreated) were used for RNA extraction and 10 μg of RNA was loaded per lane. (B) Comparison of MeJA-induced VSP1 and LOX2 expression between 35S:ANAC019-1# and wild-type plants. (C) RNA gel blot analysis showing enhanced expression of ANAC055 in different ANAC055-overexpressing lines. Tissues of 2-week-old seedlings (untreated) were used for RNA extraction and 10 μg of RNA was loaded per lane. (D) Comparison of MeJA-induced VSP1 and LOX2 expression between 35S:ANAC055-4# and wild-type plants. For (B and D), 2-week-old plants were treated with 50 μM MeJA and tissues were collected at the indicated times for RNA extraction. In all, 10 μg of total RNA was loaded per lane. Duplicated gels stained with EtBr were used as loading controls.
In the absence or presence of a range of concentrations of MeJA, 35S:ANAC019-1# and 35S:ANAC055-4# seedlings showed a root phenotype that was indistinguishable from that of wild-type plants (Figure 5C). The two transgenic lines were then compared with the wild type for MeJA-induced defense gene expression. As shown in Figure 6B and 6D, RNA gel blot analysis indicates that the overexpression of ANAC019 or ANAC055 does not lead to constitutive expression of VSP1 and LOX2. However, the MeJA-induced expression levels of these transcripts were found to be significantly higher in the transgenic lines than in the wild-type plants. Together, these results demonstrate that the overexpression of ANAC019 or ANAC055 enhances the responses of plants to MeJA in defense gene expression.
Overexpression of ANAC019 partially rescues alterations of MeJA-induced defense gene expression in the atmyc2-2 mutant
To study the action sites of the two NAC genes in the JA signaling, ANAC019 was overexpressed in the genetic background of the JA-insensitive mutant atmyc2-2 (Figure 7A). It has previously been shown that AtMYC2 differentially regulates two branches of JA-induced defensive genes and, therefore, the atmyc2-2 mutant exhibits decreased expression of JA-induced VSP1 versus elevated expression of JA-induced PDF1.2 19, 25. In agreement with this, our results indicate that the overexpression of ANAC019 in the atmyc2-2 background does not lead to the constant expression of VSP1 or PDF1.2 in the absence of MeJA; it merely rescues, at least partially, the alterations of MeJA-induced VSP1 and PDF1.2 expression of the atmyc2-2 mutant (Figure 7B). These results suggest that constitutive expression of ANAC019 bypasses AtMYC2 for JA-induced defense gene expression and support the idea that ANAC019 probably acts downstream of AtMYC2 in the JA signaling. Further evidence to support this idea came from the finding that the reduction of MeJA-induced VSP1 expression in anac019 anac055 was not as severe as in atmyc2-2 (Figure 7C).
Overexpression of ANAC019 in the coi1-1 and atmyc2-2 mutant backgrounds. (A) RNA gel blot analysis of ANAC019 expression in the various backgrounds as indicated. In all, 10 μg of total RNA from 2-week-old untreated plants was loaded per lane. A duplicated gel stained with EtBr was used as a loading control. (B) RNA gel blot analysis of MeJA-induced VSP1 and PDF1.2 expression in different genotypes. (C) RNA gel blot analysis of MeJA-induced VSP1 expression in the indicated genotypes. (D) RNA gel blot analysis of MeJA-induced VSP1 expression in wild-type, coi1-1 and coi1-1/35S:ANAC019 transgenic plants. For B to D, 2-week-old plants were treated with 50 μM MeJA for 6 h and tissues were collected for RNA extraction. In all, 10 μg of total RNA was loaded per lane. A duplicated gel stained with EtBr was used as a loading control.
Given that elegant studies have demonstrated that AtMYC2 functions downstream of COI1 to regulate JA-induced defense gene expression 19, 25, it is reasonable to predict that the action site of ANAC019 localizes downstream of COI1 and AtMYC2. To test this, we crossed the above-described 35S:ANAC019-1# with coi1-1 and identified plants that had overexpression of ANAC019 in the genetic background of coi1-1 (Figure 7A). However, overexpression of ANAC019 failed to rescue the MeJA-induced VSP1 expression of the coi1-1 mutant (Figure 7D), suggesting that the function of COI1 is still required for MeJA-induced VSP1 expression even in transgenic plants that constitutively express ANAC019. This result is consistent with the fact that COI1 is required for the sensitivity of plants to JA. Supporting evidence to this idea came from the recent finding that the SCFCOI1-JAZ complex might be the site of JA-Ile perception 22, 23.
Knockout or overexpression of the two NAC genes affects plant responses to pathogen attack
Given the established roles of JA in regulating plant defense responses against pathogens, especially necrotrophic pathogens 3, 39, we were interested in finding out whether the two NAC genes are involved in these processes. Indeed, our RNA gel blot analysis indicated that the expression of ANAC019 and ANAC055 was induced by pathogen inoculation (Figure 8A). We then examined the performance of anac019 anac055, 35S:ANAC019-1# and 35S:ANAC055-4# against a necrotrophic fungus, Botrytis cinerea. As a control, wild-type plants and the atmyc2-2 mutant were also included in these experiments. Four-week-old plants were challenged with B. cinerea and symptoms of infection were scored. In agreement with previous studies, atmyc2-2 showed an increased resistance to this pathogen. Like atmyc2-2, the anac019 anac055 double mutant also showed increased resistance to B. cinerea compared with the wild type, as indicated by fewer symptoms of infection in the infected plants (Figure 8B). By contrast, transgenic lines overexpressing ANAC019 or ANAC055 showed decreased resistance to this pathogen (Figure 8B).
Knockout or overexpression of ANAC019 and ANAC055 affects plant responses to B. cinerea. (A) B. cinerea infection induced the expression of ANAC019 and ANAC055 in wild-type plants. Wild-type plants were inoculated with 5 × 105 spores ml−1 of B. cinerea. Leaves were collected for RNA extraction 2 days after inoculation. In all, 30 μg of total RNA was loaded per lane. (B) Graphical representation of disease symptoms 5 days after inoculation of the leaves infected with 5 × 105 spores ml−1 of B. cinerea. The results shown are representative of three independent experiments.
Discussion
Our data indicate that ANAC019 and ANAC055 have an important role in regulating JA-signaled defense responses. First, the expression of the two NAC genes was induced by MeJA in a COI1- and AtMYC2-dependent manner (Figure 1). Second, an in vitro protein-DNA-binding assay showed that ANAC019 interacts with a cis-element (CATGTCCACG) in the promoter region of VSP1 (Figure 2). Third, the anac019 anac055 double mutant plants showed attenuated MeJA-induced VSP1 and LOX2 expression (Figure 5D), whereas transgenic plants overexpressing ANAC019 or ANAC055 showed enhanced MeJA-induced VSP1 and LOX2 expression (Figure 6B–6D). Fourth, overexpression of ANAC019 partially rescued the JA-related phenotype of atmyc2-2 (Figure 7B), which disrupted the function of the bHLH transcription factor AtMYC2, a recently identified player involved in JA-signaling in Arabidopsis 19, 25. Fifth, the susceptibility of the anac019 anac055 double mutant to a necrotrophic fungus showed high similarity to that of the atmyc2-2 mutant (Figure 8). Together, these data led us to a hypothesis that the two NAC proteins might act downstream of AtMYC2 to regulate JA-mediated defense responses.
An interesting aspect of the role of the two NAC transcription factors in JA-signaling concerns the phenotypes of the anac019 anac055 double mutant, which showed decreased expression of MeJA-induced VSP1 and LOX2 (Figure 5D), but increased resistance to the necrotrophic pathogen B. cinerea (Figure 8). This observation is consistent with those from mutants with disrupted AtMYC2 function. Previous studies have showed that, in response to JA treatment, AtMYC2 promotes the expression of a group of wound response-related genes, whereas it represses the expression of a group of pathogen defense-related genes. Consistently, knockout mutations in AtMYC2 have shown a reduced expression of VSP1 and LOX2 versus an increased expression of pathogen defensive PDF1.2 and PR-1 and, as a consequence, have exhibited a significant increase in their resistance to the necrotrophic pathogen B. cinerea 19, 25. Similarity of mutant phenotype, together with our finding that overexpression of ANAC019 (in the genetic background of atmyc2-2) rescues the alterations caused by MeJA-induced VSP1 and PDF1.2 expression in the atmyc2-2 mutant (Figure 7B), provides supporting evidence that ANAC019 and ANAC055 act downstream of AtMYC2 in the JA signaling.
Further evidence to support the idea that ANAC019 acts downstream of AtMYC2 came from a comparison of MeJA-induced expression levels of VSP1 among 35S:ANAC019-1#, anac019 anac055 and atmyc2-2. As shown in Figure 7C, in contrast to 35S:ANAC019-1#, which showed higher expression levels of VSP1 compared with the wild type, both anac019 anac055 and atmyc2-2 showed reduced expression levels of VSP1 compared with the wild type. However, the reduction of MeJA-induced VSP1 expression in anac019 anac055 was not as severe as that in atmyc2-2.
In contrast to the reported alleles of the atmyc2 mutant, which are less sensitive than the wild type to the inhibition effect of JA on root growth 19, 25, our root-growth assay indicates that the anac019 anac055 double mutant does not show an apparent root phenotype in the presence of MeJA (Figure 5C). This may be explained by the existence of other ANAC-related proteins; for example, ANAC072 (At4g27410) shows high sequence similarity to ANAC019 and ANAC055. Further possibilities rely on the existence of unknown components (branches), which may function in parallel with the ANAC proteins to regulate JA-mediated root growth.
Materials and Methods
Plant materials and growth conditions
All A. thaliana lines used were on the Columbia (Col-0) background. The anac019 (SALK_096295) and anac055 (SALK_014331) mutants were obtained from the Arabidopsis Biological Resources Center. The JA response mutants coi1-1 14 and atmyc2-2 have previously been described 25.
Arabidopsis seeds were surface-sterilized with 30% bleach and 0.001% Triton X-100 for 10 min and washed three times with sterile water. Sterilized seeds were then suspended in 0.2% agarose and plated on Murashige and Skoog medium. Plants were vernalized in darkness for 3 d at 4 °C and then transferred to a phytotrone set at 22 °C with a 16-h light/8-h dark cycle. After 2-3 weeks, seedlings were also potted in soil and placed in a growth room at 22 °C with a 16-h light/8-h dark cycle.
Identification of T-DNA insertion lines and generation of the anac019 anac055 double mutant
Putative knockout mutant lines of anac019 (SALK_096295) and anac055 (SALK_014331) were identified from the SALK T-DNA insertion library database (http://signal.salk.edu/cgi-bin/tdnaexpress). Seeds were obtained from the Arabidopsis Biological Resource Center (ABRC) (Ohio State University), and seedlings were analyzed individually using PCR amplification to confirm the presence of T-DNA using the LBa1 primer (located in the T-DNA) and gene-specific primers (Supplementary information, Table S1). A SALK_096295 plant that was homozygous for the T-DNA insertion was further backcrossed to the wild type and the resulting F2 progeny were germinated on kanamycin-containing medium. An F2 population consisting of 251 individuals was scored, and the segregation ratio of kanamycin-resistant versus kanamycin-sensitive seedlings was found to be 192:59 (which is close to 3:1). These results suggest that a single T-DNA insertion was present in the homozygous SALK_096295 plant. Kanamycin-resistant F2 seedlings were subjected to another round of PCR analysis to identify plants that were homozygous for the T-DNA insertion. F3 progeny of the identified SALK_096295 plants that were homozygous for the T-DNA insertion were then examined by RNA gel blot for a lack of ANAC019 expression in the presence or absence of MeJA. Based on these analyses, we identified a homozygous T-DNA insertion line with disrupted expression of ANAC019 (designated as anac019). Using a similar procedure, we also identified a homozygous T-DNA insertion line with disrupted expression of ANAC055 (designated as anac055).
anac019 and anac055 were crossed and an anac019 anac055 double mutant line was identified from the resulting F2 population by PCR analysis. Linkage analysis indicated that the JA-related phenotype of the double mutant was co-segregated with the double T-DNA insertions (data not shown).
Transgenic manipulation of ANAC019 or ANAC055 expression in wild-type (Col-0), atmyc2-2 and coi1-1 backgrounds
The coding sequences of ANAC019 and ANAC055 were amplified by RT-PCR and cloned into the BamHI and SacI sites of the binary vector pBI121 under the control of the 35S promoter of cauliflower mosaic virus. The primers that were used are listed in Supplementary information, Table S1. The resulting 35S:ANAC019 and 35S:ANAC055 constructs were introduced into wild-type Arabidopsis plants using Agrobacterium tumefaciens-mediated transformation 40. T2 seeds from each of the selected transgenic plants were plated on germination medium containing kanamycin as selection antibiotics, and the homozygous lines were selected. Homozygous T3 progeny were then examined for the expression levels of the target genes by RNA gel blot analysis. In total, 12 homozygous lines showing elevated expression of ANAC019 and 7 lines showing elevated expression of ANAC055 were identified. Representative lines overexpressing ANAC019 (Figure 6A) or ANAC055 (Figure 6C) were used for further analysis. Results from ANAC019-1# and ANAC055-4# are shown in this report.
To get ANAC019-overexpressing plants in the atmyc2-2 background, the above-mentioned ANAC019 coding sequence was cloned into the BamHI and SacI sites of the pCAMBIA1300-221-HA vector and the resulting plasmid was introduced into atmyc2-2 plants. Selection of positive transformants was conducted on germination medium containing hygromycin as an antibiotic selection marker. T3 progeny of transgenic lines that were homozygous for the ANAC019 transgene were subjected to RNA gel blot analysis for elevated levels of ANAC019 expression (Figure 7A) and PCR analysis for the presence of the atmyc2-2 allele 25.
To get ANAC019-overexpressing plants in the coi1-1 background, 35S:ANAC019-1#, which showed increased expression of ANAC019 (Figure 6A), was crossed to the coi1-1 mutant and the resulting F1 plants were selfed. Kanamycin-resistant F2 seedlings were analyzed individually with a cleaved amplified polymorphic sequence (CAPS) marker 14 to confirm the presence of the coi1-1 mutation. The identified plants were further examined using northern blot techniques to verify the overexpression of ANAC019 (Figure 7A).
Physiological assays
To carry out a root growth inhibition assay, Arabidopsis seeds were germinated on MS medium containing specific concentrations of MeJA (Sigma, St Louis, MO) and the root length was scored after 7 days of growth on vertical plates. To carry out a JA-induced defense gene expression assay, 2-week-old seedlings that were grown on MS medium were sprayed evenly with solutions containing specific concentrations of MeJA and then incubated in a growth chamber under continuous light. Tissues were harvested at specific time intervals for RNA extraction.
Plant infection with pathogens
Four-week-old Arabidopsis plants were used for pathogen inoculation based on published methods 25 with minor modifications. For inoculation with B. cinerea, fungal progression and infection symptoms were monitored for 10 days, and infection ratings from 0 to 3 were assigned to the inoculated plants (0, no infection/necrosis; 1, leaves showing some necrosis; 2, leaves showing severe necrosis; 3, dead/decayed leaves), based on a previously described method 25. In each experiment, at least 15 plants per genotype were inoculated and 4 leaves from each plant were scored for symptom development. Experiments were repeated at least three times with similar results.
RNA gel blot analysis
Total RNA extraction and northern blot analysis were conducted according to a published method 37. RNA gel blots were probed with PCR-amplified DNA fragments using gene-specific primers (Supplementary information, Table S1).
Subcellular localization of ANAC019 and ANAC055
The cDNA of ANAC019 was amplified by PCR, digested by SalI and SpeI, and fused in frame with GFP to a pBA-GFP vector, which contains a 35S promoter. The PCR-amplified cDNA of ANAC055 was digested with SpeI and fused in frame with GFP to the pBA-GFP vector. After sequencing confirmation, the 35S:ANAC019-GFP and 35S:ANAC055-GFP constructs, as well as the 35S:GFP vector control, were introduced into wild-type plants as described above and homozygous T3 transgenic plants were obtained based on antibiotic selection. The root tips of 7-day-old transgenic seedlings were visualized with a laser scanning confocal microscope (LSM 510, Zeiss, Oberkochen, Germany).
Yeast assays
Yeast assays were conducted using the previously described yeast strain HF7c, which contains the two reporter genes His3 and LacZ 32. To test the dimerization of the two NAC proteins, the PCR-derived full-length coding sequences of ANAC019 and ANAC055 were cloned into the pGBKT7 vector containing the GAL4 DNA-binding domain (pBD), and the pGADT7 vector containing the GAL4 activation domain (pAD). The resulting plasmids were used in a yeast two-hybrid assay 32. To test the transcription activation activity of ANAC019 and ANAC055, their C-terminal fragments (amino acids 148-317), named ANAC019C and ANAC055C, respectively, were cloned into the pGBKT7 vector (pBD) and transformed into yeast. The transcription activation activity of each protein was evaluated based on a published protocol 32. The primers that were used are shown in Supplementary information, Table S1.
Production of His-ANAC019 fusion proteins and in vitro DNA-binding assay
The pET28a vector (Novagen) was used to construct a plasmid expressing His-tagged fusions of ANAC019. The primers that were used are shown in Supplementary information, Table S1. Production and purification of the His-tag fusion protein was performed according to the manufacturer's instructions. The wild-type and mutant oligonucleotide vsp probes used (see legend of Figure 2) were designed according to the 10-bp element 'CATGTCCACG', which presents in the promoter region of VSP1. Oligonucleotide probes were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (Pharmacia). The DNA-binding assays were performed in the absence or presence of a non-radiolabeled competitor at room temperature in a final volume of 20 μL with a binding buffer of 15 mM Hepes, pH 7.5, 35 mM KCl, 1 mM EDTA, 6% glycerol, 1 mM DTT, 1 mM MgCl2, and 2 μg of poly(dI-dC). The samples were incubated at room temperature for 30 min and then separated on 2.0% agarose gel. After drying, the gels were autoradiographed.
(Supplementary Information is linked to the online version of the paper on the Cell Research website.)
References
Li L, Li C, Lee GI, Howe GA . Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc Natl Acad Sci USA 2002; 99:6416–6421.
Ryan CA, Moura DS . Systemic wound signaling in plants: a new perception. Proc Natl Acad Sci USA 2002; 99:6519–6520.
Turner JG, Ellis C, Devoto A . The jasmonate signal pathway. Plant Cell 2002; 14(Suppl):S153–S164.
Stratmann JW . Long distance run in the wound response—jasmonic acid is pulling ahead. Trends Plant Sci 2003; 8:247–250.
Howe GA . Jasmonates as signals in the wound response. J Plant Growth Regul 2004; 23:223–237.
Schilmiller AL, Howe GA . Systemic signaling in the wound response. Curr Opin Plant Biol 2005; 8:369–377.
Mason HS, Mullet JE . Expression of two soybean vegetative storage protein genes during development and in response to water deficit, wounding, and jasmonic acid. Plant Cell 1990; 2:569–579.
Hildmann T, Ebneth M, Pena-Cortes H, Sanchez-Serrano JJ, Willmitzer L, Prat S . General roles of abscisic and jasmonic acids in gene activation as a result of mechanical wounding. Plant Cell 1992; 4:1157–1170.
Penninckx IA, Thomma BP, Buchala A, Metraux JP, Broekaert WF . Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 1998; 10:2103–2113.
Overmyer K, Tuominen H, Kettunen R, et al. Ozone-sensitive arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 2000; 12:1849–1862.
Rao MV, Lee H, Creelman RA, Mullet JE, Davis KR . Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 2000; 12:1633–1646.
Kong HY, Jung HW, Lee SC, Choi D, Hwang BK . A gene encoding stellacyanin is induced in Capsicum annuum by pathogens, methyl jasmonate, abscisic acid, wounding, drought and salt stress. Physiol Plant 2002; 115:550–562.
Feys B, Benedetti CE, Penfold CN, Turner JG . Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 1994; 6:751–759.
Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG . COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998; 280:1091–1094.
Devoto A, Nieto-Rostro M, Xie D, et al. COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J 2002; 32:457–466.
Xu L, Liu F, Lechner E, et al. The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 2002; 14:1919–1935.
Staswick PE, Su W, Howell SH . Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 1992; 89:6837–6840.
Berger S, Bell E, Mullet JE . Two methyl jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol 1996; 111:525–531.
Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R . JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004; 16:1938–1950.
Staswick PE, Tiryaki I, Rowe ML . Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 2002; 14:1405–1415.
Dombrecht B, Xue GP, Sprague SJ, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 2007; 19:2225–2245.
Chini A, Fonseca S, Fernandez G, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007; 448:666–671.
Thines B, Katsir L, Melotto M, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007; 448:661–665.
Anderson JP, Badruzsaufari E, Schenk PM, et al. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 2004; 16:3460–3479.
Boter M, Ruiz-Rivero O, Abdeen A, Prat S . Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 2004; 18:1577–1591.
Bell E, Creelman RA, Mullet JE . A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc Natl Acad Sci USA 1995; 92:8675–8679.
Penninckx IA, Eggermont K, Terras FR, et al. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 1996; 8:2309–2323.
Olsen AN, Ernst HA, Leggio LL, Skriver K . NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 2005; 10:79–87.
Souer E, van Houwelingen A, Kloos D, Mol J, Koes R . The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996; 85:159–170.
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M . Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 1997; 9:841–857.
Aida M, Ishida T, Tasaka M . Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 1999; 126:1563–1570.
Xie Q, Frugis G, Colgan D, Chua NH . Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 2000; 14:3024–3036.
Tran LS, Nakashima K, Sakuma Y, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004; 16:2481–2498.
He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY . AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 2005; 44:903–916.
Delessert C, Kazan K, Wilson IW, et al. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J 2005; 43:745–757.
Ooka H, Satoh K, Doi K, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 2003; 10:239–247.
Zheng W, Zhai Q, Sun J, et al. Bestatin, an inhibitor of aminopeptidases, provides a chemical genetics approach to dissect jasmonate signaling in Arabidopsis. Plant Physiol 2006; 141:1400–1413.
Utsugi S, Sakamoto W, Murata M, Motoyoshi F . Arabidopsis thaliana vegetative storage protein (VSP) genes: gene organization and tissue-specific expression. Plant Mol Biol 1998; 38:565–576.
Glazebrook J . Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 2005; 43:205–227.
Bechtold N, Pelletier G . In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 1998; 82:259–266.
Acknowledgements
We are grateful to Dr Xinnian Dong (Duke University, Durham, NC, USA) for critical reading of the manuscript and valuable suggestions. We thank Dr Jianmin Zhou (National Institute of Biological Sciences, Beijing, China) for providing the fungus strain Botrytis cinerea, Dr Salomé Prat (Institut de Biologia Molecular de Barcelona, Barcelona, Spain) for providing homozygous atmyc2-2 (T-DNA insertion line SALK_083483) seeds and Dr Daoxin Xie (Tsinghua University, Beijing, China) for providing the coi1-1 seeds. This work was supported by grants from The National Natural Science Foundation of China (30530440), The Ministry of Science and Technology of China (2006CB102004, 2006AA10A116), and The Chinese Academy of Sciences (KSCX2-YW-N-045).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary information Table S2
Oligonucleotide primers used in this study (PDF 36 kb)
Rights and permissions
About this article
Cite this article
Bu, Q., Jiang, H., Li, CB. et al. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res 18, 756–767 (2008). https://doi.org/10.1038/cr.2008.53
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2008.53
Keywords
This article is cited by
-
OsNAC103, an NAC transcription factor negatively regulates plant height in rice
Planta (2024)
-
Transcriptome analysis of sugarcane reveals rapid defense response of SES208 to Xanthomonas albilineans in early infection
BMC Plant Biology (2023)
-
Genome-wide analyses of the NAC transcription factor family to reveal the potential candidate genes responding to powdery mildew in balsam pear
Plant Biotechnology Reports (2023)
-
Comparative transcriptome analysis of resistant and susceptible wheat in response to Rhizoctonia cerealis
BMC Plant Biology (2022)
-
PtoNF-YC9-SRMT-PtoRD26 module regulates the high saline tolerance of a triploid poplar
Genome Biology (2022)