Abstract
Salmonella can invade non-phagocytic cells through its type III secretion system (T3SS-1), which induces a Trigger entry process. This study showed that Salmonella enterica, subspecies enterica serovar Enteritidis can also invade cells via the Rck outer membrane protein. Rck was necessary and sufficient to enable non-invasive E. coli and Rck-coated beads to adhere to and invade different cells. Internalization analysis of latex beads coated with different Rck peptides showed that the peptide containing amino acids 140-150 promoted adhesion, whereas amino acids between 150 and 159 modulated invasion. Expression of dominant-negative derivatives and use of specific inhibitors demonstrated the crucial role of small GTPases Rac1 and Cdc42 in activating the Arp2/3 complex to trigger formation of actin-rich accumulation, leading to Rck-dependent internalization. Finally, scanning and transmission electron microscopy with Rck-coated beads and E. coli expressing Rck revealed microvillus-like extensions that formed a Zipper-like structure, engulfing the adherent beads and bacteria. Overall, our results provide new insights into the Salmonella T3SS-independent invasion mechanisms and strongly suggest that Rck induces a Zipper-like entry mechanism. Consequently, Salmonella seems to be the first bacterium found to be able to induce both Zipper and Trigger mechanisms to invade host cells.
Similar content being viewed by others
Introduction
Bacteria have evolved various mechanisms to trigger rearrangements of host actin, enabling their entry into the host cells. In some cases, bacteria express surface proteins that interact with cellular transmembrane receptors connected to the cytoskeleton. This type of entry has been described for Yersinia and Listeria invasion as 'Zipper mechanism' 1. In other cases, such as Salmonella and Shigella, the bacteria bypass the interaction with a membrane receptor and deliver bacterial effector proteins directly into the cytosol via a type III secretion system (T3SS) ('Trigger mechanism'). These two bacterial entry mechanisms target small Rho guanosine triphosphatase proteins (RhoGTPases) and consequently manipulate host cell architecture to induce transient actin rearrangement 2. For example, a subset of small RhoGTPases and the conserved actin nucleator, Arp2/3 complex, are regulated during invasin-stimulated internalization of Yersinia 3.
Salmonella are Gram-negative facultative bacteria that target a variety of eukaryotic hosts, causing a broad spectrum of diseases from gastroenteritis to typhoid fever in humans and animals. To infect their hosts, Salmonella have evolved mechanisms to interact with non-phagocytic cells, to induce their own internalization and to survive within the host environment. The genetic basis for Salmonella invasion is complex, and numerous bacterial genes necessary for Salmonella internalization have been identified. The main Salmonella internalization mechanism, and the one that has been studied most extensively, requires the T3SS-1. This T3SS, encoded by the Salmonella pathogenicity island 1 (SPI-1), is a molecular syringe that injects bacterial effector proteins directly into host cells to manipulate host cell pathways, allowing bacterial invasion through a Trigger mechanism 4. Another T3SS, encoded by the SPI-2, allows intracellular survival and multiplication of Salmonella within the vacuole 5.
Other entry mechanisms involving PagN and Rck have been described in Salmonella 6, 7. PagN is a 26 kDa outer membrane protein (OMP) mediating adhesion to and invasion of epithelial cells by interacting with heparinated proteoglycan 8. Rck is a 19-kDa OMP encoded by the rck gene on the large virulence plasmid of the bacteria 6. In addition to adhesion and entry capabilities, Rck also confers a high level of resistance to the bactericidal activity of complement, independent of the lipopolysaccharide (LPS) structure, by inhibiting the polymerization of complement component C9 in the bacterial membrane 9, 10. Rck belongs to a family of five homologous OMP proteins characterized in Salmonella (Rck and PagC), Escherichia coli (Lom), Yersinia enterocolitica (Ail) and Enterobacter cloacae (OmpX) 11. Molecular analysis has shown that Rck is homologous to PagC and Ail with 53% and 48% identity, respectively, but complement resistance and invasion have been attributed only to Rck and Ail 11. However, the precise role of Rck in Salmonella invasion process is still unknown, as well as the mechanism of the Rck-dependent internalization pathway. The aim of this study was to verify the fact that Rck is a factor for invasion, to identify its role in Salmonella and to analyze the signaling pathway induced within host cells.
Results
Rck confers invasiveness to non-invasive bacteria
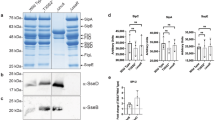
To clearly demonstrate that Rck is an OMP involved in eukaryotic cell adhesion and invasion, we overexpressed Rck in a non-invasive E. coli strain (BL21 pLysS). To address this question, we first tested different constructs (His-Rck and GST-Rck). As we obtained the best invasion level with E. coli BL21 pLysS transformed with pGEX-4T-2 Rck (E. coli-rck) after IPTG induction, we performed a subcellular fractionation analysis 12, 13. The predominant localization of mature Rck (Rck without its signal peptide) in the OmpA-enriched fraction corresponding to the outer membrane proves that Rck is expressed at the membrane level (Figure 1A). The cleavage of the fusion protein at the junction between GST and the fusion partner has already been described in the recombinant protein handbook: protein amplification and simple purification (18-1142-75, Amersham pharmacia biotech). Rck was also detected in the cytosol and the inner membrane fractions, but in lower quantities. The majority of GST-Rck was, in contrast, within inclusion body fractions. To demonstrate that Rck at the membrane level is active, E. coli-rck was also tested for its ability to resist complement-mediated killing. It was observed that E. coli BL21 pLysS transformed with the vector alone (E. coli-GST) suffered more than 6 logs of killing and did not survive in pooled human serum. In contrast, E. coli-rck showed a high level of resistance to killing, since less than 1 log of bacteria were killed by the complement.
Rck is able to induce adhesion and internalization of a non-invasive E. coli strain. (A) Comparison of the partitioning of Rck in subcellular fractions. Two consecutive fractions were collected as described in Materials and Methods and analyzed by western blotting with specific antibodies to Rck, Hsp60, OmpA and SecG (IB, inclusion body fractions; OM, outer membrane fractions; IM, inner membrane fractions; C, cytoplasmic fractions). Adhesion (B) and invasion (C) of epithelial cells by Rck. HT29 (gray bars) and MA104 cells (hatched bars) were infected with E. coli-GST and E. coli-rck (MOI, 1:5) for 1 h at 37 °C as indicated. The percentage of adherent (B) and internalized bacteria (C) was determined as described in Materials and Methods. (D) Invasion inhibition by soluble Rck. MA104 cells were infected with E. coli-rck in the presence of purified Rck-His proteins as indicated. Treatment with different concentrations of BSA in elution buffer had no effect. Thus, only the highest concentration (15 μg BSA) is shown. The number of adherent (white bars) and internalized bacteria (black bars) was determined and expressed relative to values obtained for untreated cells (CTL), arbitrarily set at 100%. Data are the mean ± SD of at least three independent experiments, with two infected wells evaluated per experiment. Data were compared using a Student's t-test. **P < 0.01, ***P < 0.001.
Induction of Rck in recombinant E. coli-rck conferred this strain with the ability to bind to and invade epithelial (MA104, HT29, Figure 1B and 1C), fibroblastic (NIH-3T3, data not shown), trophoblastic and endothelial (Jeg-3 and HBrMEC, Supplementary information, Figure S1) cell lines. The highest invasion level was obtained with MA104 cells, in which the invasion rate of the E. coli-rck strain was 10 000-fold higher than that of the control (E. coli-GST) (Figure 1B and 1C). The lowest level was obtained with HT29 cells with only a 10-fold higher invasion rate by E. coli-rck than by the control E. coli-GST (Figure 1B and 1C). The other cell lines tested were intermediary, as invasion was 100-fold higher in the NIH-3T3, Jeg-3 and HBrMEC cells by the E. coli-rck than by the mock-transformed E. coli (Supplementary information, Figure S1).
To demonstrate that Rck did not have an indirect effect on adhesion and invasion by modifying either the E. coli membrane surface or the gene regulation, the Rck-mediated internalization was inhibited with different concentrations (from 0.1 to 15 μg) of purified Rck protein tagged with (His)6 (lacking its signal peptide (Rck-His)) (Figure 1D). The results showed that the percentage of internalized E. coli-rck decreased drastically in the presence of Rck-His and in a dose-dependent manner compared to control experiments. Overall, these data confirm the results of Heffernan et al. 6, showing that Rck is able to mediate both adhesion and invasion. Because MA104 cells allowed the highest invasion level, the remaining experiments were shown only with this cell line.
Rck contributes to Salmonella internalization
To assess further the role of Rck OMP in Salmonella, a Δrck mutant (LA5 Δrck) was constructed. Adhesion and invasion assays showed that rck deletion did not significantly reduce adhesion to and entry of Salmonella Enteritidis into MA104 and NIH-3T3 cell lines (Supplementary information, Figure S2A). This result could be related to the fact that Rck was not expressed in our in vitro culture conditions. To investigate this, we first analyzed the transcription of endogenous rck by quantitative reverse transcription-PCR (RT-PCR) in Salmonella at different growth phases. It was observed that rck was expressed at a very low level, notably at the stationary growth phase of bacteria used for cell infection and also in exponential phase (data not shown). To bypass the low in vitro rck expression, Rck was overexpressed in the wild-type and the Δrck strains by transformation with the multi-copy plasmid pGEX-4T-2 carrying rck. Adhesion and invasion assays performed with bacteria at the end of their exponential phase showed that Rck overexpression significantly increased invasion of MA104 and NIH-3T3 cells (P < 0.01), but had no statistically significant effect on bacterial adhesion (Figure 2A and Supplementary information, Figure S2B). The low in vitro expression of Rck in the wild-type strain was therefore probably related to the fact that the transcription of this gene is regulated by quorum sensing, which involves SdiA in an N-acyl homoserine lactone (AHL)-dependent manner 14. Adhesion and invasion assays were therefore performed with bacteria cultivated in the presence of AHL and under a swarming condition known to activate cell-cell signaling systems 15. Under these conditions, rck deletion induced a significant (P < 0.01) decrease in invasion, resulting in more than 50% less invasion than the wild-type strain (Figure 2B). No effect was observed on adhesion, probably due to the presence of numerous other bacterial factors on the Salmonella strain. This effect was related to Rck, as complementation with a pUC19 vector carrying rck restored the wild-type invasion phenotype. These data indicate that Rck contributes to Salmonella invasion, for at least some cell lines and/or cell types in addition to T3SS-1.
Rck contributes to Salmonella invasion. (A) Adhesion and invasion of the wild-type (wt) S. Enteritidis LA5 strain overexpressing GST (white bars) or GST-Rck (gray bars), an rck-deleted LA5 strain overexpressing GST (black bars) or GST-Rck (hatched bars). Strains were grown in TSB and then deposited for 1 h at 37 °C onto MA104 cells. Results are expressed relative to values obtained for LA5 GST (arbitrarily set at 100%). (B) Adhesion and invasion of the LA5 wild-type strain (white), the rck-deleted strain (black) or the rck-deleted strain complemented with pUC19-rck (hatched). Strains were grown under swarming conditions and then deposited onto MA104 cells. Results are expressed relative to values obtained for the LA5 strain (arbitrarily set at 100%). Five independent experiments were performed per cell line, with at least two infected wells evaluated for each experiment. All data are shown as average percentages of attached or internalized bacteria ± standard error of the mean (SEM). Data were compared using a Welch test. **P < 0.01, ***P < 0.001.
Rck-triggered uptake is preceded by formation of an actin-rich accumulation
By using a recombinant non-adherent and non-invasive E. coli BL21 pLysS strain, the activity of Rck can be assayed without the influence of other Salmonella invasion proteins. To investigate the hypothesis that E. coli-rck invades epithelial cells by hijacking cellular actin, the organization of actin microfilaments within MA104 cells during invasion by E. coli-rck was analyzed using confocal microscopy. F-actin was labelled in red using phalloidin conjugated to rhodamin (Figure 3A). Permeabilized bacteria were localized with an anti-GST antibody labelled in green. Rck-expressing bacteria were observed associated with MA104 cells (Figure 3B and 3C). Stacks of confocal images were generated and analyzed by z sectioning using Imaris software (Bitplane) (Figure 3C). These sections showed a localization of polymerized actin encircling individual and small clusters of bacteria.
Rck is able to bind to cells and to locally remodel the actin cytoskeleton. (A-C) MA104 cells were infected with E. coli-rck (MOI 1:5) for 30 min at 37 °C and then processed for immunofluorescence. Confocal laser scanning microscopy shows horizontal (A-B) and vertical sections of cells (C). (A): actin staining in red. (B-C): overlay of actin in red and GST in green, yellow areas indicate co-localization of actin and GST. A-C are representative images with an arrow indicating the site of local E. coli-rck-induced actin polymerization and typical structural morphologies, more clearly visible in the insets (A-C, bar = 5 μm; in insets bar = 2 μm). (D): Effect of cytochalasin D on Rck-mediated adhesion to cells and invasion. The ability to adhere to (white bars) and invade cells (black bars) was assessed in the presence of different concentrations of cytochalasin D. Results are expressed relative to values obtained for the same amount of DMSO containing medium, arbitrarily set at 100%. Each value represents the mean ± SD of at least three independent experiments, with two infected wells evaluated per experiment. Data were compared using a Student's t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
To assess the role of actin filament polymerization in Rck-mediated invasion, the effect of cytochalasin D was analyzed. Cytochalasin D is a fungal metabolite that binds to the barbed end of actin filaments, thus inhibiting the association and dissociation of actin monomers at that end 16. Before adhesion and invasion assays, increasing concentrations of cytochalasin D were added to cell monolayers, resulting in a dose-dependent decrease in the number of internalized bacteria compared to control cells. A 17-fold decrease in invasion was observed with 0.5 μg/ml cytochalasin D (Figure 3D). In contrast, the addition of cytochalasin D over the same concentration range resulted in no significant changes in the total number of cell-associated bacteria, providing further evidence that adhesion and internalization are discrete events and that only the latter is dependent on actin microfilament function.
A 46 amino acid region of Rck is essential for cellular binding and internalization
As demonstrated above, the adhesion of Rck-expressing bacteria to MA104 cells leads to their invasion, a process that might require energy derived from the metabolic activity of the microbe and other compounds present in the E. coli strain. To demonstrate that Rck alone can induce cell invasion, GST-Rck fusion protein was isolated from E. coli-rck and coated onto 2 μm latex beads. The fact that these beads attached to and were internalized by MA104 cells in the same manner as E. coli-rck demonstrated that Rck is an invasin. As a control, beads were coated with GST alone. Adhesion of extracellular beads to the MA104 cells, detected by their double fluorescence due to their green autofluorescence and red-labelled antibodies against GST, demonstrates that Rck can mediate adhesion onto cells. Moreover, intracellular Rck-coated beads were characterized by their autofluorescence alone, demonstrating that Rck induces internalization of beads. This entry was specific, as beads coated with GST alone were rarely associated with cells and never found within cells (data not shown). Moreover, as observed with E. coli-rck, internalization of inert particles coated with GST-Rck was also mediated by actin polymerization (Table 1). Stacks of confocal images analyzed by z-sectioning using Imaging software also showed that beads had indeed been internalized (data not shown).
To identify the functional domain in Rck, a mutagenesis strategy was undertaken, according to the previously proposed structural model 17. Six deletions in the loop 3 and/or 4 of the GST-Rck fusion protein were constructed: 72 (RckΔ113), 45 (RckΔ140), 35 (RckΔ150) and 26 (RckΔ159) amino acids of the Rck C-terminal domain were deleted, and peptides containing the amino acids between 113 and 185 (Rck113-185) and between 113 and 159 (Rck113-159) were engineered (Figure 4A). Because some of these Rck peptides were not well expressed on the bacterial surface as GST fusion proteins, they were purified from E. coli BL21 pLysS and coated onto 2 μm latex beads. Table 1 shows that the amino acids between 1 and 113 (loop 1 and 2) and between 159 and 185 (loop 4) were not involved in adhesion or entry mechanisms. On the other hand, three mutants (RckΔ159, 113-185 Rck and 113-159 Rck) showed adhesion, actin recruitment (labelled with phalloidin) and entry abilities similar to the wild-type protein (Table 1, Figure 4B-4D and Supplementary information, Figure S3). As mutant RckΔ140 showed diminished adhesion and mutant RckΔ150 exhibited adhesion but not invasive properties (Figure 4E), we hypothesized that peptide 140-159 might be involved in adhesion, whereas peptide 150-159 might be involved in internalization. To investigate this hypothesis further, GST-Rck peptides 140-159 and 150-159 were engineered. They were however unable to mediate adhesion or invasion, suggesting that they were unable to adopt an appropriate conformation. Interestingly, the ability of Rck mutants to mediate internalization was correlated with their ability to trigger F-actin accumulation at sites of particle binding, whereas mutants that only mediated adhesion did not recruit actin (Table 1, Figure 4 and Supplementary information, Figure S3).
The 113-159 peptide of Rck is able to bind to cells and to locally remodel the actin cytoskeleton. (A) Different E. coli BL21 pLysS strains expressing GST-RckΔ113, GST-RckΔ140, GST-RckΔ150, GST-RckΔ159, GST-113-185 Rck, GST-113-159 Rck, GST-140-159 Rck and GST-150-159 Rck were engineered. MA104 cells were incubated with (B-D) GST-113-159 Rck or (E) GST-RckΔ150-coated beads at 37 °C for 30 min and stained by immunofluorescence. Confocal laser scanning microscopy shows horizontal (B, C and E) and vertical sections of cells (D). (B) Actin staining in red. (C, D and E) overlay of actin and beads in green; yellow areas indicate co-localization of actin and beads. B-E are representative images with an arrow indicating the site of local bead-induced actin polymerization and typical structural morphologies, more clearly visible in the insets (B, C and E, bar = 20 μm; D, bar = 5 μm; and in insets bar = 2 μm).
To demonstrate that peptide 113-159 contains the active invasion site, the next step was therefore to test the ability of GST-113-159 Rck-coated beads to inhibit the entry of E. coli-rck into cultured epithelial cells. We observed in Figure 5 that GST-113-159 Rck-coated beads induced an increase of E. coli-rck adhesion to cells, due to Rck autoagglutination. The 113-159 peptide seems to be involved in internalization, because only 7% of the adherent E. coli-rck are internalized. In contrast, GST-coated beads did not modify either adhesion or internalization capabilities of E. coli-rck. Similarly, GST-RckΔ150-coated beads, which have a lower adhesion capability than GST-113-159 Rck-coated beads and no internalization capability, induced a slight decrease in E. coli-rck adhesion and 45% of adherent bacteria were internalized.
Rck-mediated invasion is inhibited by 113-159 Rck peptides. Cells were incubated with 57 × 104 GST, GST-113-159 Rck or GST-RckΔ150-coated beads per well for 15 min at 4 °C prior to infection with E. coli-rck for 1 h at 37 °C. The ability of the bacteria to adhere to (white bars) and invade cells (black bars) was assessed and expressed relative to values obtained for untreated cells (CTL), arbitrarily set at 100%. Each value represents the mean ± SD of at least three independent experiments, with two infected wells per experiment. Data were compared using a Student's t-test. **P < 0.01, ***P < 0.001.
The interactions of E. coli-rck or the GST-113-159 Rck-coated beads with MA104 (Figure 6) or Jeg-3 (Supplementary information, Figure S4) cell surface were further analyzed by scanning and transmission electron microscopy. Different stages of invasion can be visualized: adherent beads associated with cellular membrane via microvillus-like extensions, partially engulfed beads or bacteria with a weak membrane rearrangement (Figure 6A-6C and Supplementary information, Figure S4A-S4D) and totally internalized beads or bacteria (Figure 6A and 6C; Supplementary information, Figure S4A). Rck-dependent membrane rearrangements are not as marked as for a Trigger mechanism, and are similar to the membrane engulfment described for Listeria invasion 2, 18. This observation suggests that Rck mediates a Zipper-like internalization process. Because actin accumulation could be visualized more clearly with GST-113-159 Rck-coated beads, these particles were used for the remaining experiments.
The Arp2/3 complex is involved in Rck-mediated internalization
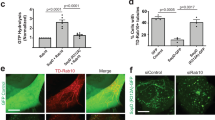
A nucleator molecule is needed to initiate efficient actin filament formation and elongation. The Arp2/3 complex is a highly conserved actin nucleator, composed of two actin-related proteins (Arp2 and Arp3) and five other unrelated proteins. The Arp2/3 complex is able to nucleate de novo actin filaments and elongate them at their barbed ends, allowing internalization of bacteria 19, 20. To investigate the pathway leading to actin-mediated internalization, double fluorescence staining was performed using an antibody against Arp3 and rhodamine-phalloidin. Figure 7A showed that Arp 3 was recruited at the Rck-coated beads entry site, 30 min after incubation. Moreover, these experiments indicated the co-localization of actin and Arp3 during Rck-induced internalization.
The Arp2/3 complex is necessary for Rck-dependent internalization. (A) Recruitment of Arp3 during entry of GST-113-159 Rck-coated beads in MA104 cells. Arp3 (in green) and actin (in red) colocalize at the entry sites (white arrow) of the beads (GST staining in blue), more clearly visible in the insets (bar = 10 μm; in insets bar = 2 μm). (B) Effect of the overexpression of Scar WA on actin polymerization. MA104 cells were transfected for 18 h with Scar WA-myc, incubated with GST-113-159 Rck-coated beads for 30 min at 37 °C and then processed for confocal microscopy to visualize Scar WA-myc (in green), actin (in red) and GST (in blue). A and B are representative images, with an arrow indicating typical staining patterns, more clearly visible in the insets. (C) Quantification of actin recruitment to adherent particles per cell. The adhering particle per cell (white bars) and actin recruitment per particle (black bars) are expressed relative to the values obtained for untransfected cells. Data were compared using a Student's t-test. ***P < 0.001.
To obtain clear evidence that the Arp2/3 complex regulates the internalization process, we analyzed the actin accumulation induced by Rck-coated beads in MA104 cells transfected with Scar WA, corresponding to the C-terminal WASP-homology, and acidic domains of Scar1, an Arp2/3 complex activator 21. This truncated protein binds the Arp2/3 complex, and its overexpression sequesters Arp2/3 in the cytosol. Figure 7B and 7C showed that the actin-rich accumulation around Rck-coated beads in Scar WA-transfected cells reduced drastically. Overall, these results indicate that the actin nucleation activity of the Arp2/3 complex is required for Salmonella Rck-mediated actin polymerization.
Small GTPases Rac and Cdc42 are involved in Rck-mediated internalization
The involvement of Arp2/3 complex-dependent actin polymerization led us to investigate the role of the small RhoGTPases in Rck-mediated internalization. To determine whether the RhoGTPases are differentially recruited to the contact site surrounding GST-113-159 Rck-coated beads, MA104 cells were transfected with the Myc- or GFP-tagged versions of the wild type form of GTPases. Fluorescence microscopy showed that internalized GST-113-159 Rck-coated beads were associated with F-actin. Expression of tagged versions of RhoA, Rac or Cdc42 revealed that only Rac and Cdc42 were recruited with actin around the beads (Figure 8A-8C).
Rac and Cdc42, but not Rho, are involved in Rck-dependent internalization. MA104 cells were transfected for 18 h with GFP Cdc42 wild type (wt) (bar = 10 μm) (A), myc Rac wt (bar = 20 μm) (B) or GFP Rho wt (bar = 20 μm) (C), and incubated with GST-113-159 Rck-coated beads for 30 min at 37 °C. They were processed for confocal microscopy to visualize actin in red, GFP or myc in green and beads in blue. (A-C) Representative images with an arrow indicating typical staining patterns, more clearly visible in the insets (in insets bar = 2 μm). (D) Before infection with E.coli-rck (MOI 1:5), MA104 cells were pre-treated for 90 min with different concentrations of TAT-N17Rac, TAT-N17Cdc42 or 4 h with TAT-C3-transferase. The percentage of cell-associated bacteria and internalized bacteria was determined 1 h after infection. The binding (white bars) and internalization (black bars) indices were related to the control values obtained with the same amount of desalting medium (CTL), arbitrarily set at 100%. Error bars represent the standard deviation of the mean values obtained from three different experiments with two infected wells evaluated per experiment. Data were compared using a Student's t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
TAT fusion proteins allow transduction of proteins into all cells in culture, and allow the level of expression and concentration of the respective proteins to be monitored over time. In this study, the TAT-myc protein was fused with the dominant-negative form of Rac (N17Rac) and Cdc42 (N17Cdc42) that were produced in E. coli. First, the ability of TAT fusion proteins to transduce into MA104 cells was studied. A kinetic analysis of the amount of TAT-fusion proteins within cells determined by western blot analysis showed that 90 min of incubation allowed the maximum level of TAT-mediated transduction into cells (data not shown). The transduced TAT fusion proteins were active, since microspike formation was blocked by the TAT-N17Cdc42 protein after stimulation with bradykinin (Supplementary information, Figure S5A) 22. Moreover, stimulation of Rac-dependent ruffles with sphingosine-1-phosphate was inhibited by transduction of the TAT-N17Rac protein (Supplementary information, Figure S5B) 22. These tools allowed the role of Rac and Cdc42 in Rck-triggered invasion to be analyzed. Under our conditions, none of the dominant-negative form of Rac and Cdc42, incubated before infection, had any substantial effect on the adhesion of E. coli-rck onto MA104 cells. In contrast, they led to almost complete inhibition of Rck-mediated internalization (Figure 8D). Further experiments revealed that a recombinant cell-permeable form of C3 toxin (TAT-C3), an ADP-ribosylating protein of Clostridium botulinum that specifically inhibits RhoA, B and C 23, showed no inhibitory effect on the adhesion and invasion of E. coli-rck (Figure 8D). Thus, Rck-mediated entry appears to be mediated by a combination of Rac and Cdc42, but not Rho.
Discussion
To infect different hosts and cause various diseases ranging from typhoid fever to gastroenteritis or to lead to an asymptomatic carrier state, Salmonella are able to cross several barriers. Crossing these barriers and multiplying within the host require invasion of a large variety of phagocytic and non-phagocytic cells. Most of researches have focused on understanding the T3SS-1-dependent entry mechanism. However, Salmonella express other molecules that allow them to invade host cells. Two OMPs have been described as being involved in this entry process, namely, PagN and Rck 6, 7. However, their respective role in Salmonella pathogenesis is poorly understood, as is their internalization mechanism. The pagN gene is activated by PhoP and the corresponding protein shares identities with the Hek and Tia invasins/adhesins of pathogenic E. coli 7. The Rck protein is encoded by different serovars, including Salmonella Enteritidis and Salmonella Typhimurium, and is a member of a 17- to 19-kDa OMP family involved in cell adhesion and/or invasion including Ail (Yersinia enterocolitica), PagC (Salmonella Typhimurium), OmpX (Enterobacter cloacae) and Lom (bacteriophage lambda-lysogenic E. coli) 11.
Heffernan et al. 6 have previously shown that Rck overexpressed in a non-invasive E. coli strain enables the bacteria to adhere to and invade fibroblastic cells. However, the role of Rck in Salmonella was not demonstrated. Our study confirmed that Rck expressed in a non-invasive E. coli strain confers the ability to adhere to and invade epithelial, fibroblastic and endothelial cells. We also demonstrated that the purified recombinant protein coated on beads was necessary and sufficient to promote adhesion to and invasion of cells contrary to beads coated with GST alone. In Salmonella, rck is not expressed in vitro in standard culture conditions, but is induced by quorum sensing and especially by SdiA 24. Consequently, we observed that under conditions known to induce quorum sensing, that is, swarming conditions 15, the lack of rck in Salmonella Enteritidis led to a significant decrease in invasion compared to the wild-type strain. On the other hand, overexpression of Rck in Salmonella increased the entry capability of Salmonella. These results demonstrate that Rck plays a role in Salmonella invasion.
The fact that Rck is regulated by quorum sensing suggests an intestinal role of this protein. Quorum sensing is known to induce virulence genes in numerous bacteria, such as enteropathogenic E. coli, and especially in the intestine 25. In Salmonella Typhimurium, it was shown that SdiA is activated in the gastrointestinal tract of mice infected with Yersinia enterocolitica and of turtles infected with Aeromonas hydrophila 26, 27. Interestingly, Rck seems to play a role in the mouse intestine since a Salmonella Typhimurium strain expressing AHLs from Yersinia, which is more competitive than an SdiA mutant in mice, loses its fitness advantages when rck is deleted. Moreover as rck is also regulated by an unidentified SdiA-independent system at 37 °C and 42 °C, it is conceivable that Rck invasion mechanism is not restricted to the gastrointestinal tract 14. These data and the fact that Rck allows a weak level of entry into the human intestinal epithelial cells (HT29) could suggest that Rck exhibits host and/or organ specificity. A better understanding of the conditions controlling rck expression should help to further analyze the in vivo role of Rck.
Currently, no crystal structure is available for Rck, but in the proposed structural model, Rck is predicted to be an eight-stranded β-barrel protein that resides in the outer membrane 17. Use of deletion mutants suggests that residues 140-150 of Rck are responsible for adhesion to MA104 cells, whereas peptide 150-159 is involved in internalization. However, this study did not allow adhesion and internalization to be induced with the peptide 140–159. As the 113-159 peptide induces both adhesion and entry of coated beads, we hypothesize that peptide 113-140 confers functional conformation to the 140-159 peptide. This hypothesis is supported by Cirillo et al. 17 who showed that G118D mutation greatly reduced Rck-mediated cell invasion. In fact, we observed that this single amino acid substitution inhibited adhesion and thus cell internalization (Supplementary information, Figure S6). Ultrastructure analysis will enable this issue to be investigated further.
The scanning and transmission electron microscopy analysis of the interaction between either GST-113-159 Rck-coated beads or E. coli-rck and epithelial cell surface showed a Zipper-like structure engulfing the adherent coated beads or bacteria within a vacuole. These data, combined with the fact that Rck alone mediates internalization and that soluble Rck inhibited the entry mediated by Rck, strongly suggest that Rck mediates cell invasion through a Zipper-like mechanism, as previously described for other bacteria such as Listeria and Yersinia 2, 18. This is the first report to show that Salmonella can enter cells through a Zipper-like mechanism in addition to the Trigger mechanism mediated by its T3SS-1 apparatus. The prototype of Zipper-like bacterial internalization is that of Yersinia, which involves the subversion of the β1 integrin in a receptor-mediated mechanism 2. The receptor-ligand interaction promotes the microfilament cytoskeleton-dependent advance of the pseudopod and receptor clustering, leading to a signal transduction through focal adhesion kinase and the stimulation of cytoskeletal rearrangements by small GTP-binding proteins 2.
Rho family GTPases are differentially targeted by intracellular pathogens to promote invasion. For bacteria like group B Streptococcus and Vibrio parahaemolyticus, all three Rac, Cdc42 and Rho GTPases are involved in cell invasion 28, 29. In contrast, Chlamydia trachomatis requires solely Rac for internalization 30. In other cases, a combination of two GTPases allows invasion, as in the case of Candida albicans that needs Rho and Rac to modulate actin cytoskeleton organization 31. Specific inhibitors and expression of dominant-negative derivatives demonstrated that the Rck-mediated entry process was dependent on the mobilization of actin microfilaments that were activated by the signaling Arp2/3 complex and the GTPases Rac and Cdc42, but not by Rho. The role of these signaling molecules is however not specific to the Zipper mechanism, as their role has also been described in Trigger entry processes 32, 33, 34. For example, the T3SS-1 effector SopE activates Cdc42 and Rac by mimicking the action of eukaryotic G-nucleotide exchange factors 35, remodeling the cytoskeleton and altering gene expression during Salmonella T3SS-1-dependent invasion. Then, Cdc42 activates members of the WASP family, whereas Scar/WAVE is activated by Rac during Salmonella T3SS-1-dependent invasion. Recent data suggested that WAVE complex induces ruffling but is dispensable for efficient entry, whereas a novel Arp2/3 complex activator, WASH, mediates entry 36. Activation of the Arp2/3 complex, which initiates actin nucleation and branching, results in membrane ruffling and/or bacterial uptake depending on the Arp2/3 complex activator 37. Hence, regulation of Rac- and Cdc42-mediated signaling events seems to be crucial for Salmonella to successfully establish infection through a Zipper and a Trigger entry process. However, it is far from clear how these molecules contribute to Salmonella invasion. Salmonella could provide an interesting model to investigate this question, since, to our knowledge, it is the first bacterium found to be able to induce both a Trigger and a Zipper-like mechanism to invade cells. Yersinia spp. use indeed its T3SS to inhibit the host immune system through a diverse array of mechanisms including inhibition of phagocytosis by disrupting the actin cytoskeleton and induction of apoptosis in macrophages. Investigation of the molecular elements of the signal transduction and regulation pathway mediated by Rck and T3SS-1 will allow a better understanding of the Salmonella invasion process and could help to explain the specificity of each invasion pathway.
Materials and Methods
Reagents
Cytochalasin D (Sigma) was dissolved in dimethyl sulfoxide (DMSO) at a stock concentration of 1 mg/ml. The maximum final concentration of DMSO never exceeded 0.1% (v/v) in drug-treated cells.
Cell lines and transfection
HBrMEC were human brain microvascular endothelial cells provided by C Kieda (CNRS UPR, Orleans, France) 38. HBrMEC were cultured in OptiMEM-1 with Glutamax (Invitrogen) supplemented with inactivated 2% fetal bovine serum (Invitrogen) and antibiotics (penicillin 100 IU/ml and streptomycin 100 μg/ml). African green monkey fetal kidney epithelial cells (MA104, HPACC: 85102918) and human Caucasian colon adenocarcinoma cells (HT29, ATCC: HTB-38) were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 25 mM glucose supplemented with inactivated 10% fetal bovine serum 2 mM, L-glutamine and antibiotics. NIH 3T3 mouse fibroblasts (ATCC: CRL-1658) were cultured in DMEM containing 25 mM glucose supplemented with inactivated 10% fetal bovine serum, 2 mM L-glutamine, 1mM sodium pyruvate and antibiotics. Jeg-3 cells (human epithelial placental cells ATCC no: HTB-36) were grown in MEM medium containing Glutamax, non-essential aminoacids, sodium pyruvate and 10% fetal bovine serum. For transfection, cells were seeded onto coverslips and transfected as previously described 39. A total of 1 μg DNA was used to transfect 3 × 104 cells on a 24-well plate, using SuperFect (Qiagen) as recommended by the manufacturer. After 3 h, fresh medium was added for 16 h and the cells were then antibiotic starved.
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in Table 2. Bacteria were routinely grown in Luria-Bertani (LB) broth with shaking at 150 rpm at 37 °C overnight. Salmonella LA5 wild-type strain, isolated from infected chickens 40, Δrck mutant and pUC19-rck complemented Δrck strains were cultured under swarming conditions as described by Kim and Surette 15. Briefly, overnight cultures in nutrient broth (NB) were diluted in the ratio 1:10 000 in fresh NB, and 100 μl were spread-plated on swarm NB with 0.5% Glucose (NBG) and 0.5% agar. After 5 h at 37 °C, 200 nmol of N-hexanoyl-homoserine lactone (Fluka) were added on filter disks deposited on the plate surface 7 h before infection assays. Strains were routinely grown in LB broth or in TSB with the corresponding antibiotics kanamycin (Km) 25 or 50 μg/ml, nalidixic acid (Nal) 20 μg/ml, chloramphenycol (Cm) 34 μg/ml and carbenicillin (Cb) 100 μg/ml. Before induction of Rck expression, overnight cultures were diluted (1:20) in LB.
DNA constructs
Eukaryotic expression vector (pRK5) encoding myc-tagged wild-type Rho, Rac, N17Rac and N17Cdc42 have been described previously 39, 41. pRK5-Myc Scar WA was provided by P Cossart (Institut Pasteur, Paris). The plasmid of pTAT-C3 was provided by J Bertoglio (INSERM U 461, Faculté de pharmacie Paris Sud) and it was introduced into E. coli strain BL21 pLysS 42. GFP-Cdc42 was generated by subcloning wild-type Cdc42 from pRK5-myc Cdc42 into pEGFP-C2 (CLONTECH Laboratories Inc; BamHI and EcoRI restriction sites). The rck mutant of LA5 strain was obtained using the lambda-Red mutagenesis system, modified as previously described 43, 44. The sequences of P1 and P2 primers used for this strain construction are listed in Table 3. The rck gene was amplified from Salmonella Enteritidis LA5 wild-type strain by PCR using the sense primer rck forw (flanked by BamHI restriction site) and the reverse primer rck rev (flanked by EcoRI restriction site) or with the reverse primer rck-P3 (flanked by HindIII) and cloned into pGEX-4T-2 or pUC19 expression vector, respectively (Amersham-Pharmacia), before being transformed into E. coli BL21 pLysS. The same method was used to construct the rck without its peptide signal (Rck-His) into pET15b using primers rck-His forw and rck-His rev, flanked by NdeI and XhoI restriction sites, respectively.
Rck-deleted mutants were engineered by PCR by introducing a premature stop codon at positions 114 (RckΔ113), 141 (RckΔ140), 151 (RckΔ150) and 160 (RckΔ159), using pGEX-4T-2 Rck as a template, with the sense primer rck forw, flanked by BamHI restriction site, and the following antisense primers rck Δ113 rev for rck Δ113, rck Δ140 rev for RckΔ140, rck Δ150 rev for RckΔ150 and rck Δ159 rev for RckΔ159, flanked by EcoRI restriction site. The 113-185 Rck mutant was engineered by PCR, using pGEX-4T-2 Rck as a template, with the sense primer 113-185 rck forw, flanked by BamHI restriction site, and rck rev as antisense primer, flanked by EcoRI restriction site. The 113-159 Rck mutant was engineered by PCR, using pGEX-4T-2 RckΔ159 as a template, with the sense primer 113-185 rck forw, flanked by BamHI restriction site, and rck Δ159 rev as antisense primer, flanked by EcoRI restriction site. The 150-159 Rck mutant was engineered by PCR, using pGEX-4T-2 RckΔ159 as a template, with the sense primer rck Δ150 forw, flanked by BamHI restriction site, and PstI rev as antisense primer, flanked by PstI restriction site. The 140-159 Rck mutant was engineered by PCR, using pGEX-4T-2 RckΔ159 as a template, with the sense primer rck Δ140 forw, flanked by BamHI restriction site, and PstI rev as antisense primer, flanked by PstI restriction site. The amplified DNA fragments, digested with BamHI and EcoRI or with BamHI and PstI, were cloned into pGEX-4T-2 expression vector before being transformed into E. coli BL21 pLysS. The G118D Rck mutant was first engineered by PCR using pGEX-4T-2 113-185 Rck as a template, the sense primer rck lp3 G118D forw, flanked by BamHI restriction site, and the antisense primer rck rev. The amplified DNA was inserted in pGEX-4T-2 and digested with KpnI and XhoI. Then, the remaining 113-185 G118D Rck was subcloned in KpnI/XhoI sites of pGEX-4T-2 Rck, generating pGEX-4T-2 G118D Rck.
A sequence coding for the TAT peptide (GYGRKKRRQRRRGGFLN), flanked by the EagI and XhoI restriction sites, was cloned downstream of the (His)6 coding sequence in pET-23a-d(+). The new vector is referred to as pET-(His)6-TAT vector. Care was taken to make use of the codon usage for bacteria. PCR-amplified coding sequences of myc-N17Cdc42, flanked by HindIII and SalI, were then subcloned in HindIII/XhoI sites of pET-(His)6-TAT vector. The myc-N17Rac coding sequence, flanked by the NdeI and XmaI restriction sites, was also subcloned in pET-(His)6-TAT vector using the same restriction sites, before being transformed into E. coli BL21 pLysS. Thus, the tags (His, TAT and Myc, in this order) were at the N-terminal of the protein leaving the C-terminal accessible for acylation. All constructs were validated by sequencing.
Fractionation procedure
An overnight culture of BL21 E. coli-rck was diluted in 200 ml LB broth and GST-Rck expression was induced with 1 mM IPTG. After 4 h of induction, bacteria were centrifuged for 10 min at 4800× g. Pellet was resuspended in 10 ml LB and centrifugated for 10 min at 2 000× g. Pellet was resuspended in 1.6 ml HEPES (10 mM; pH 7.4) containing 20% sucrose. Bacterial lysis was performed by sonication and unbroken bacteria were removed by a centrifugation for 20 min at 2 000× g. The membrane fraction was separated from the cytoplasm as previously described by Ishidate et al. 12. The inner and outer membrane of the inclusion bodies were separated using a second sucrose gradient, as described by Schnaitman 13.
Quantitative RT-PCR
Reverse transcription-PCR was performed to evaluate the relative transcriptional level of rck gene compared to the housekeeping gene tufA. RNAs were extracted from exponential and stationary growth phase cultures of LA5 wild-type strain. RNA extraction, DNase treatment and quantitative RT-PCR were performed as previously described 43. rck and tufA RNAs were amplified with rck-R1 and tufA-A5 sense primers and rck-R2 and tufA-A6 antisense primers, respectively.
Serum sensitivity studies
Normal human serum was collected from healthy volunteers, pooled and stored at −80 °C until use (pooled normal human sera). Control sera were decomplemented by heating at 56 °C for 30 min (heat-inactivated sera). Serial 10-fold dilution of mid-log-phase bacterial suspension in phosphate-buffered saline (PBS) were incubated for 60 min in 50% pooled normal human or heat-inactivated sera at 37 °C and then plated on Tryptic Soy Agar (TSA). Serum sensitivity was calculated as the difference between CFUs surviving incubation in pooled normal human sera and those in heat-inactivated sera, expressed in log base 10 and designated as log kill 6.
Expression and purification of fusion proteins
Protein expression was induced in subcultures of E. coli BL21 pLysS expressing various Rck constructs in pGEX-4T-2 with 1 mM IPTG for 4 h at 37 °C. For protein purification, cells were harvested by centrifugation, resuspended in buffer containing 50 mM Tris (pH 8), 40 mM EDTA, 25% sucrose, 100 mM MgCl2, 0.2% Triton X-100, 1 mM phenylmethylsulfonylfluoride and complete protease inhibitor cocktail (Boehringer) and sonicated. After clearing, fusion proteins were affinity purified from the soluble fraction on Glutathione-Sepharose 4B beads (Amersham Biosciences) according to the manufacturer's instructions. Rck-His expression was induced in subcultures of E. coli BL21 pLysS with 1 mM IPTG for 2 h at 37 °C. Bacteria were harvested by centrifugation, and protein purification was performed under denaturing conditions using QIAexpress NI-NTA Fast Start kit (Qiagen). Purified proteins were stored at −80 °C.
(His)6-TAT N17Rac and (His)6-TAT N17Cdc42 protein expression were induced in subcultures of E. coli BL21 pLysS with 1mM IPTG for 3 h at 27 °C. Bacteria were pelleted and resuspended in lysis buffer (pH 8) containing 100 mM NaH2PO4, 10 mM Tris, 8 M urea, 0.05% Tween 20, 10 mM imidazole and 10 mM β-mercaptoethanol. After sonication, the lysate was centrifuged and the supernatant was then incubated at 4 °C overnight with NiNTA agarose beads. Beads were washed with the same buffer as above but at pH 6.5. Elution was performed with the same buffer at pH 4.5. Proteins were stored at 4 °C. To eliminate urea, proteins were passed through a Biogel PD10 column (Biorad) equilibrated in DMEM supplemented with 500 mM NaCl. The desalting procedure diluted the proteins by two-fold. Time between the desalting procedure and addition of the protein to the cells was <5 min. Controls were prepared by adding the same amount of NaCl-DMEM-containing medium. (His)6-TAT-C3 proteins were expressed by IPTG induction (1 mM IPTG, 4 h, 25 °C). Recombinant (His)6-TAT-C3 was extracted from E. coli BL21 pLysS strain using QIAexpress Ni-NTA fast start kit under native conditions. The column was washed with a 10 mM imidazole buffer solution and eluted using a 250 mM imidazole elution buffer. The purity of each TAT-protein preparation was determined on polyacrylamide gels stained with Coomassie blue.
Adhesion and invasion assay
Cell monolayers were grown in 24-well tissue culture plates (Falcon) for 3-4 days to obtain a confluent monolayer. Before infection, cells were incubated overnight in medium without antibiotics. They were infected for 60 min at 37 °C with 300 μl of bacterial suspension in cell culture medium without fetal calf serum, at a bacterium-to-cell ratio of 5:1 for E. coli strains or 10:1 for bacteria grown under swarming conditions. For adhesion assays, after bacteria-cell contact, cells were gently washed at least four times with PBS and then disrupted with 1 ml cold distilled water. Viable bacteria (intra- and extra-cellular) were counted after plating serial dilutions on TSA. Invasion was quantified by the gentamicin protection assay to kill remaining extracellular bacteria, as previously described 45. After incubation with 100 μg/ml gentamicin, cells were washed and then lysed by adding 1 ml cold distilled water. The number of viable bacteria released from the cells was counted as for adhesion assays.
Rck-His inhibition assay
Samples of 0.1, 10 and 15 μg of purified Rck-His or BSA were incubated with MA104 cells in 0.2 ml of DMEM in 24 wells simultaneously with bacterial suspension. Plates were incubated for 60 min at 37 °C and the infection assay was performed as described above. Controls were performed with BSA in elution buffer, diluted in culture medium as for Rck-His.
Coating of latex beads with proteins and peptides
About 109 latex beads (2 μm diameter, polystyrene sulfate modified, Sigma) were washed and resuspended in PBS. Purified GST, GST-Rck and the different GST-Rck-peptide fusion proteins (1 mg) were added and adsorbed onto the beads for 3 h at room temperature. After adding BSA (20 mg/ml), the solution was incubated at room temperature for another hour. The beads were then washed in PBS containing 1 mg/ml BSA and stored at 4 °C. To determine the coupling efficiency, the protein concentration of the starting solution and the supernatant were determined before adding BSA. In addition, coated GST-Rck fusion proteins were checked using western blot analysis.
SDS-PAGE and western blot analysis
All GST-Rck fusion protein-coated beads were denaturated in SDS sample buffer (about 2.5 × 106 beads were loaded per lane). Separation of the proteins was performed using SDS-PAGE. For immunoblotting, proteins were transferred electrophoretically onto polyvinylene difluoride (PVDF) membranes (1.2 mA/cm2 for 60 min). Sheets were blocked for 1 h with PBS containing 5% nonfat dry milk at room temperature. For detection of GST fusion proteins, a polyclonal rabbit anti-GST (Sigma; dilution 1: 2 000) and a peroxydase-conjugated secondary anti-rabbit antibody (Amersham, diluted in the ratio 1:10 000) were added for 1 h each. The detection was carried out using a chemiluminescence detection kit (ECL) from Amersham. A partitioning of Rck in subcellular E.coli-rck fractions was undertaken. Each fraction was examined by western blotting with anti-OmpA given by Roland Lloubes (diluted in the ratio 1:3 000), anti-SecG antibody provided by Professor Koreaki Ito (Kyoto University, Kyoto, Japan; diluted in the ratio 1:10 000), anti-Hsp60 (Stressgen, diluted in the ratio 1:2 500) and anti-Rck (diluted in the ratio 1:2 500). A kinetic analysis of the amount of TAT-fusion proteins within cells was determined. Cell extracts were separated by 15% SDS-PAGE and revealed by western blotting with anti-myc antibodies (Santa Cruz, diluted in the ratio 1:500).
Immunofluorescence microscopy
Cell monolayers on coverslips were incubated overnight in cell culture medium without antibiotics before addition of beads (about 150 beads per cell). After incubation at 37 °C in a humidified atmosphere at 5% CO2 for 30 min in an appropriate medium without fetal calf serum, cells were washed in PBS to remove unbound beads. Cells were fixed for 10 min in 4% paraformaldehyde solution and permeabilized for 5 min in 0.2% Triton-X 100 in PBS. Actin was stained with Rhodamin-Phalloidin (Sigma; diluted in the ratio 1:100). Actin nucleator Arp2/3 complex was stained with a polyclonal antibody recognizing Arp3 (Millipore, diluted in the ratio 1:100), myc with antibody 9E10 (Santa Cruz, diluted in the ratio 1:100) and GST with a polyclonal antibody (Sigma, diluted in the ratio 1:100). Secondary antibodies were Alexa 488, Alexa 568, Alexa 647-labelled goat anti-mouse or anti-rabbit (Molecular probes; diluted in the ratio 1:200 each). Finally, coverslips were mounted in fluorescence mounting medium (Dako) and analyzed with an Olympus Fluoview 500 confocal laser-scanning microscope (Olympus). Different staining procedures helped to differentiate internalized from total associated GST-Rck-coated latex beads. As all latex beads were coated with GST-Rck, cells were incubated with rabbit anti-GST antibody (diluted 1:100). Cells were washed with PBS to remove unbound antibody and incubated with alexa 568-labelled goat antibody anti-rabbit antibodies. In this way, internalized beads (green due to the fluorescence of beads) were distinguished from extracellular beads (yellow due to an overlay of red and green fluorescence), providing a means to score particle invasion. Finally, coverslips were mounted in fluorescence mounting medium and analyzed with the confocal laser-scanning microscope.
Scanning electron microscopy
For scanning electron microscopy analysis, cells on coverslips, incubated for 1 h with GST-113-159 Rck-coated beads or E. coli-rck, were washed in PBS and then fixed in a mixture of 4% paraformaldehyde and 1% glutaraldehyde in PBS (0.3 M; pH 7.4) for 1 h. The cells were washed overnight with PBS (pH 7.4) and dehydrated through a graded series of 50%, 70%, 90% and 100% acetone solution. Then, they were dried to CO2 critical point and coated with a thin layer of platinum on a Jeol JUC 5000 sputtering coater. Samples were observed on Gemini 982 Zeiss FEGSEM.
Transmission electron microscopy
For transmission electron microscopy analysis, cells incubated with GST-113-159 Rck-coated beads or E. coli-rck were scraped off with a Cell Scraper (Falcon), washed in PBS, pelleted and fixed by incubation for 48 h in 4% paraformaldehyde and 1% glutaraldhehyde in 0.1 M phosphate buffer (pH 7.2). Cell pellets were then washed in PBS, postfixed for 1 h with 1% osmium tetraoxide and dehydrated in a graded series of ethanol solutions and propylene oxide. Cell pellets were embedded in Epon resin (Sigma), which was allowed to polymerize for 48 h at 60 °C. Ultrathin sections deposited on 300 mesh copper grids were stained with 5% uranyl acetate and 5% lead citrate, for examination under a Jeol 1230 transmission electron microscope 46.
Statistical analysis
Data were analyzed using a parametric Student's t-test or Welch test using INSTAT v2.03 software. A Welch test was performed when the difference between the two SDs was significant.
References
Seveau S, Pizarro-Cerda J, Cossart P . Molecular mechanisms exploited by Listeria monocytogenes during host cell invasion. Microbes Infect 2007; 9:1167–1175.
Cossart P, Sansonetti PJ . Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 2004; 304:242–248.
Wiedemann A, Linder S, Grassl G, Albert M, Autenrieth I, Aepfelbacher M . Yersinia enterocolitica invasin triggers phagocytosis via beta1 integrins, CDC42Hs and WASp in macrophages. Cell Microbiol 2001; 3:693–702.
Schlumberger MC, Hardt WD . Salmonella type III secretion effectors: pulling the host cell's strings. Curr Opin Microbiol 2006; 9:46–54.
Waterman SR, Holden DW . Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol 2003; 5:501–511.
Heffernan EJ, Wu L, Louie J, Okamoto S, Fierer J, Guiney DG . Specificity of the complement resistance and cell association phenotypes encoded by the outer membrane protein genes rck from Salmonella Typhimurium and ail from Yersinia enterocolitica. Infect Immun 1994; 62:5183–5186.
Lambert MA, Smith SG . The PagN protein of Salmonella enterica serovar Typhimurium is an adhesin and invasin. BMC Microbiol 2008; 8:142.
Lambert MA, Smith SG . The PagN protein mediates invasion via interaction with proteoglycan. FEMS Microbiol Lett 2009; 297:209–216.
Heffernan EJ, Reed S, Hackett J, Fierer J, Roudier C, Guiney D . Mechanism of resistance to complement-mediated killing of bacteria encoded by the Salmonella Typhimurium virulence plasmid gene rck. J Clin Invest 1992; 90:953–964.
Taylor PW . Bactericidal and bacteriolytic activity of serum against gram-negative bacteria. Microbiol Rev 1983; 47:46–83.
Heffernan EJ, Harwood J, Fierer J, Guiney D . The Salmonella Typhimurium virulence plasmid complement resistance gene rck is homologous to a family of virulence-related outer membrane protein genes, including pagC and ail. J Bacteriol 1992; 174:84–91.
Ishidate K, Creeger ES, Zrike J, et al. Isolation of differentiated membrane domains from Escherichia coli and Salmonella Typhimurium, including a fraction containing attachment sites between the inner and outer membranes and the murein skeleton of the cell envelope. J Biol Chem 1986; 261:428–443.
Schnaitman CA . Protein composition of the cell wall and cytoplasmic membrane of Escherichia coli. J Bacteriol 1970; 104:890–901.
Smith JN, Ahmer BM . Detection of other microbial species by Salmonella: expression of the SdiA regulon. J Bacteriol 2003; 185:1357–1366.
Kim W, Surette MG . Coordinated regulation of two independent cell-cell signaling systems and swarmer differentiation in Salmonella enterica serovar Typhimurium. J Bacteriol 2006; 188:431–440.
Cooper JA . Effects of cytochalasin and phalloidin on actin. J Cell Biol 1987; 105:1473–1478.
Cirillo DM, Heffernan EJ, Wu L, Harwood J, Fierer J, Guiney DG . Identification of a domain in Rck, a product of the Salmonella Typhimurium virulence plasmid, required for both serum resistance and cell invasion. Infect Immun 1996; 64:2019–2023.
Mengaud J, Ohayon H, Gounon P, Mege RM, Cossart P . E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 1996; 84:923–932.
Millard TH, Sharp SJ, Machesky LM . Signalling to actin assembly via the WASP (Wiskott-Aldrich syndrome protein)-family proteins and the Arp2/3 complex. Biochem J 2004; 380:1–17.
Sousa S, Cabanes D, Bougneres L, et al. Src, cortactin and Arp2/3 complex are required for E-cadherin-mediated internalization of Listeria into cells. Cell Microbiol 2007; 9:2629–2643.
Machesky LM, Insall RH . Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol 1998; 8:1347–1356.
Andor A, Trulzsch K, Essler M, et al. YopE of Yersinia, a GAP for Rho GTPases, selectively modulates Rac-dependent actin structures in endothelial cells. Cell Microbiol 2001; 3:301–310.
Aktories K, Hall A . Botulinum ADP-ribosyltransferase C3: a new tool to study low molecular weight GTP-binding proteins. Trends Pharmacol Sci 1989; 10:415–418.
Ahmer BM, van Reeuwijk J, Timmers CD, Valentine PJ, Heffron F . Salmonella Typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J Bacteriol 1998; 180:1185–1193.
Sircili MP, Walters M, Trabulsi LR, Sperandio V . Modulation of enteropathogenic Escherichia coli virulence by quorum sensing. Infect Immun 2004; 72:2329–2337.
Dyszel JL, Smith JN, Lucas DE, et al. Salmonella enterica serovar Typhimurium can detect the acyl homoserine lactone production of Yersinia enterocolitica in mice. J Bacteriol 2010; 192:29–37.
Smith JN, Dyszel JL, Soares JA, et al. SdiA, an N-acylhomoserine lactone receptor, becomes active during the transit of Salmonella enterica through the gastrointestinal tract of turtles. PLoS ONE 2008; 3:e2826.
Akeda Y, Kodama T, Kashimoto T, et al. Dominant-negative Rho, Rac, and Cdc42 facilitate the invasion process of Vibrio parahaemolyticus into Caco-2 cells. Infect Immun 2002; 70:970–973.
Burnham CA, Shokoples SE, Tyrrell GJ . Rac1, RhoA, and Cdc42 participate in HeLa cell invasion by group B streptococcus. FEMS Microbiol Lett 2007; 272:8–14.
Carabeo RA, Grieshaber SS, Hasenkrug A, Dooley C, Hackstadt T . Requirement for the Rac GTPase in Chlamydia trachomatis invasion of non-phagocytic cells. Traffic 2004; 5:418–425.
Atre AN, Surve SV, Shouche YS, Joseph J, Patole MS, Deopurkar RL . Association of small Rho GTPases and actin ring formation in epithelial cells during the invasion by Candida albicans. FEMS Immunol Med Microbiol 2009; 55:74–84.
Chen LM, Hobbie S, Galan JE . Requirement of CDC42 for Salmonella-induced cytoskeletal and nuclear responses. Science 1996; 274:2115–2118.
Hardt WD, Chen LM, Schuebel KE, Bustelo XR, Galan JE . S. Typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 1998; 93:815–826.
Shibata T, Takeshima F, Chen F, Alt FW, Snapper SB . Cdc42 facilitates invasion but not the actin-based motility of Shigella. Curr Biol 2002; 12:341–345.
Friebel A, Ilchmann H, Aepfelbacher M, Ehrbar K, Machleidt W, Hardt WD . SopE and SopE2 from Salmonella Typhimurium activate different sets of RhoGTPases of the host cell. J Biol Chem 2001; 276:34035–34040.
Hanisch J, Ehinger J, Ladwein M, et al. Molecular dissection of Salmonella-induced membrane ruffling versus invasion. Cell Microbiol 2009; 12:84–98.
Unsworth KE, Way M, McNiven M, Machesky L, Holden DW . Analysis of the mechanisms of Salmonella-induced actin assembly during invasion of host cells and intracellular replication. Cell Microbiol 2004; 6:1041–1055.
Kieda C, Paprocka M, Krawczenko A, et al. New human microvascular endothelial cell lines with specific adhesion molecules phenotypes. Endothelium 2002; 9:247–261.
Wiedemann A, Patel JC, Lim J, Tsun A, van Kooyk Y, Caron E . Two distinct cytoplasmic regions of the beta2 integrin chain regulate RhoA function during phagocytosis. J Cell Biol 2006; 172:1069–1079.
Allen-Vercoe E, Dibb-Fuller M, Thorns CJ, Woodward MJ . SEF17 fimbriae are essential for the convoluted colonial morphology of Salmonella Enteritidis. FEMS Microbiol Lett 1997; 153:33–42.
Cougoule C, Hoshino S, Dart A, Lim J, Caron E . Dissociation of recruitment and activation of the small G-protein Rac during Fcgamma receptor-mediated phagocytosis. J Biol Chem 2006; 281:8756–8764.
Sebbagh M, Renvoize C, Hamelin J, Riche N, Bertoglio J, Breard J . Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat Cell Biol 2001; 3:346–352.
Fardini Y, Chettab K, Grepinet O, et al. The YfgL lipoprotein is essential for type III secretion system expression and virulence of Salmonella enterica Serovar Enteritidis. Infect Immun 2007; 75:358–370.
Figueroa-Bossi N, Ammendola S, Bossi L . Differences in gene expression levels and in enzymatic qualities account for the uneven contribution of superoxide dismutases SodCI and SodCII to pathogenicity in Salmonella enterica. Microbes Infect 2006; 8:1569–1578.
Velge P, Bottreau E, Kaeffer B, Yurdusev N, Pardon P, Van Langendonck N . Protein tyrosine kinase inhibitors block the entries of Listeria monocytogenes and Listeria ivanovii into epithelial cells. Microb Pathog 1994; 17:37–50.
Patient R, Hourioux C, Sizaret PY, Trassard S, Sureau C, Roingeard P . Hepatitis B virus subviral envelope particle morphogenesis and intracellular trafficking. J Virol 2007; 81:3842–3851.
Acknowledgements
This work was supported by the Région Centre and performed within the 'SAVIRE' project, granted by the Délégation Régionale à la Recherche et à la Technologie du Centre (FEDER) (No 1634-32245) and by the Région Centre (No 2008-00036085). M Rosselin holds a Doctoral fellowship granted by the Région Centre and the Institut National de la Recherche Agronomique. We thank C Kieda for HBrMEC cell donation, Koreaki Ito (Kyoto University, Kyoto, Japan) for anti-SecG antibody donation, P Cossart and J Bertoglio for plasmid donation and J De Rycke for his critical reading of the manuscript.
Emmanuelle Caron passed away on July 8, 2009. We would like to take the opportunity to thank her for her guidance and suppport over the years. We will miss the friend and the great scientist!
Author information
Authors and Affiliations
Corresponding author
Additional information
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Rck is able to induce adhesion to and internalization of a non invasive E. coli strain by human cells. (PDF 54 kb)
Supplementary information, Figure S2
rck deletion did not significantly reduce adhesion to and internalization of S. Enteritidis into MA104 and 3T3 cell lines. (PDF 82 kb)
Supplementary information, Figure S3
GST-113-159 Rck-coated beads are internalized by MA104 cells. (PDF 138 kb)
Supplementary information, Figure S4
Rck mediates a Zipper-like membrane rearrangement. (PDF 158 kb)
Supplementary information, Figure S5
The transduced TAT-N17Rac and -N17Cdc42 fusion proteins are active. (PDF 76 kb)
Supplementary information, Figure S6
G118D amino acid substitution in Rck inhibited adhesion and cell internalization. (PDF 37 kb)
Rights and permissions
About this article
Cite this article
Rosselin, M., Virlogeux-Payant, I., Roy, C. et al. Rck of Salmonella enterica, subspecies enterica serovar Enteritidis, mediates Zipper-like internalization. Cell Res 20, 647–664 (2010). https://doi.org/10.1038/cr.2010.45
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2010.45
Keywords
This article is cited by
-
Current Progress and Challenges in the Study of Adjuvants for Oral Vaccines
BioDrugs (2023)
-
Investigation of the invasion mechanism mediated by the outer membrane protein PagN of Salmonella Typhimurium
BMC Microbiology (2021)
-
Murine AML12 hepatocytes allow Salmonella Typhimurium T3SS1-independent invasion and intracellular fate
Scientific Reports (2021)
-
Neutralizing-antibody-mediated protection of chickens against infectious bursal disease via one-time vaccination with inactivated recombinant Lactococcus lactis expressing a fusion protein constructed from the RCK protein of Salmonella enterica and VP2 of infectious bursal disease virus
Microbial Cell Factories (2019)
-
Inhibition of Salmonella Typhimurium adhesion, invasion, and intracellular survival via treatment with methyl gallate alone and in combination with marbofloxacin
Veterinary Research (2018)