Abstract
An actinomycete strain, IR73-Li102T, was isolated from a lichen sample obtained from Iriomote Island, Japan, and subsequently characterized using a polyphasic approach. Comparative 16S rRNA gene sequence analysis revealed that strain IR73-Li102T had the highest sequence similarities with Actinomycetospora chiangmaiensis YIM 0006T (98.3%), A. corticola 014-5T (98.1%) and A. rishiriensis RI109-Li102T (98.0%). However, DNA–DNA hybridization assays, as well as physiological and biochemical analyzes, showed that strain IR73-Li102T could be clearly differentiated from its closest phylogenetic relatives. The strain contained meso-diaminopimelic acid, and arabinose and galactose were present in whole-cell hydrolysates. The predominant menaquinone was MK-8(H4), and the diagnostic phospholipids were phosphatidylethanolamine, phosphatidylcholine, phosphatidylinositol and diphosphatidylglycerol. The major cellular fatty acid was iso-C16:0 (58%). The chemotaxonomic properties of strain IR73-Li102T were consistent with those shared by members of the genus Actinomycetospora. On the basis of the phenotypic features and DNA–DNA hybridization data, strain IR73-Li102T (= NBRC 106365T = KCTC 19783T) represents a novel species of the genus Actinomycetospora, for which the name Actinomycetospora iriomotensis sp. nov. is proposed.
Similar content being viewed by others
Introduction
Lichens are frequently found in several environments, such as tree and stone surfaces and are classified into four groups based on their growth forms: foliaceous (leaf-like), fruticose (stalked), squamulose (scale-like), and crustose (crust-like).1 Biologically, lichens are symbiotic microorganisms that comprises a fungi and green algae and/or cyanobacteria.1, 2 However, several researchers have identified diverse species of actinomycetes from lichens that have the potential for producing secondary metabolites, as determined by biosynthetic gene sequences.3, 4 Recently, Motohashi et al.5 reported the isolation of a novel angucycline and butenolide from lichen-derived Streptomyces spp. Thus, evaluation of the diversity of culturable actinomycetes from lichens is an important task for understanding the ecological roles of actinomycetes and may allow the identification of novel bioactive compounds for industrial applications.
Previously, several rare actinomycetes were isolated from lichen samples collected in Japan, and were proposed as new taxa, named Nocardioides exalbidus, Leifsonia lichenia, and Actinomycetospora rishiriensis.6, 7, 8 The genus Actinomycetospora was originally proposed by Jiang et al.9 as a member of the family Pseudonocardiaceae, and was later amended by Tamura et al.10 At the time of writing, nine Actinomycetospora species have been characterized by polyphasic taxonomic approaches. The type strain of the genus Actinomycetospora, A. chiangmaiensis, was originally isolated from a soil sample in northern Thailand, whereas the other species were from soil, tree bark, and lichen samples collected in Japan.8, 9, 10
During a screening for lichen-associated actinomycetes, we isolated strain IR73-Li102T from a crustose lichen sample collected from Iriomote Island, Okinawa, Japan. The aim of the present study was to determine the taxonomic position of strain IR73-Li102T using a polyphasic taxonomic approach, which included chemotaxonomic, morphological, physiological, molecular and genomic characterizations.
Materials and methods
Isolation and maintenance of organism
A crustose lichen sample was collected from Iriomote Island, Okinawa, Japan, and was then air dried for 7 days at room temperature. The lichen sample was homogenized with mortar and pestle in sterilized distilled water, and the suspension was spread on humic acid-vitamin (HV) agar containing nalidixic acid (20 mg l−1) and cycloheximide (50 mg l−1), which was then incubated at 30 °C for 2 weeks.11 Following the incubation period, the strain IR73-Li102T was isolated and transferred to a nutrient agar (NA) plate. The strain was maintained in 20% (v/v) glycerol at −80 °C until use.
Phenotypic characterization
Strain IR73-Li102T was grown on both HV and oatmeal–YGG agar for 14 days at 28 °C, and morphological features were then analyzed by light and scanning electron microscopy (JEOL, JSM-6500F).12 Spore motility was evaluated by light microscopy using a hanging drop method with spores suspended in sterilized distilled water. Gram staining was performed by the Hucker method.13 To determine the optimal temperature and pH for growth, strain IR73-Li102T was incubated for 7 and 14 days on NA at temperatures of 5, 10, 20, 25, 30, 37 and 45 °C, and at pHs ranging from 4–12 (in 1 pH unit intervals). Growth at 5 and 10 °C was assayed after a 6-week incubation period. Growth at different NaCl concentrations (0–7%, w/v, in 1% increments) was evaluated after 14- and 21-day incubations on NA. API ZYM and API Coryne Biochemical Test Kits (bioMérieux, Tokyo, Japan) were used to evaluate several physiological and biochemical characteristics, according to the manufacturer's instructions. Assimilation of carbon sources at a final concentration of 1% (w/v) was tested using ISP medium 9 as the basal medium.14 Antimicrobial activities against selected Gram-positive and Gram-negative bacteria, yeasts and filamentous fungi were assayed using an overlay method.15 Briefly, spot-inoculated, 10-day-old colonies of strain IR73-Li102T on NA plates were overlaid with 5 ml of sloppy NA/YEPD agar inoculated with the test organisms. The sizes of the zones of inhibition around the colonies were recorded after 24 h at 30 °C.
Chemotaxonomy
Biomass for chemotaxonomic studies was obtained by growing strain IR73-Li102T in shake flasks containing glucose–yeast extract broth for 5 days at 30 °C.16 After harvesting cells by centrifugation, the resultant pellet was washed twice with distilled water. Diaminopimelic acid isomers and sugars in whole-cell hydrolysates were then analyzed on the basis of the methods established by Hasegawa et al.17 and Schaal,18 respectively. Cellular fatty acids were processed and analyzed as methyl esters, following the protocol of the MIDI Sherlock Microbial Identification System.19 Standard procedures were also used for the extraction and analysis of isoprenoid quinones and polar lipids, and the results were compared with the appropriate controls.20 The DNA G+C content of strain IR73-Li102T was determined by HPLC, as described by Tamaoka and Komagata.21
Molecular analysis
Chromosomal DNA from strain IR73-Li102T was isolated and purified by the method of Saito and Miura,22 with a minor modification.23 PCR amplification of the 16S rRNA gene from strain IR73-Li102T was performed as described by Tamura et al.,24 and the resultant PCR product was purified using a MonoFas DNA Purification Kit (GL Sciences, Tokyo, Japan). The purified PCR product was directly sequenced using an ABI Prism BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and a Genetic Analyzer automated DNA sequencer (model 3730; Applied Biosystems). The obtained 16S rRNA gene sequence was compared with published 16S rRNA gene sequences of bacterial type strains using the EzTaxon server (http://www.eztaxon.org/).25 For phylogenetic analyzes, 16S rRNA gene sequences were collected from the EMBL/GenBank/DDBJ database and aligned using the CLUSTAL_X program.26 Phylogenetic trees were constructed with the CLUSTAL_X program and Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0 using the neighbor-joining and maximum parsimony methods.26, 27, 28, 29 The PHYML software package was used to construct a maximum-likelihood tree.30 The topologies of the constructed trees were evaluated by bootstrap analysis with either 1000 (neighbor-joining and maximum-parsimony methods) or 500 resamplings (maximum-likelihood method).31
DNA–DNA hybridization
DNA–DNA hybridizations were performed as described by Kusunoki et al.,32 using biotinylated DNA, with five replications for each sample. The highest and lowest values obtained for each sample were excluded, and the mean of the remaining three values was quoted as the DNA–DNA relatedness value.
Nucleotide sequence accession number
The 16S rRNA gene sequence of strain IR73-Li102T determined in this study has been deposited in the DDBJ database under the accession number AB581531.
Results and discussion
The nearly complete 16S rRNA gene sequence (1428 nucleotide) of strain IR73-Li102T was compared with sequences of recognized bacterial species on the EzTaxon server. The highest similarity value (98.3%) was observed with A. chiangmaiensis YIM 0006T followed by A. corticola 014-5T (98.1%) and A. rishiriensis RI109-Li102T (98.0%). The 16S rRNA gene sequence similarities to members of the genus Actinomycetospora ranged from 97.0 to 98.4%.8, 9, 10 The phylogenetic tree constructed with 16S rRNA gene sequence data of members of the genus Actinomycetospora and related genera using the neighbor-joining method showed that strain IR73-Li102T formed a monophyletic clade with A. chiangmaiensis, A. corticola and A. rishiriensis (Figure 1). Moreover, this clade was strongly supported by the maximum-likelihood and maximum parsimony trees with high bootstrap values.
Phylogenetic tree derived from 16S rRNA gene sequences showing the relationship of strain IR73-Li102T to a member of the genus Actinomycetospora and related taxa. The tree was constructed using the neighbor-joining method and Knuc values.28 Only bootstrap values above 50% are shown (1000 resamplings) at the branching points. Solid circles indicate that the corresponding nodes were also recovered in maximum parsimony and maximum-likelihood algorithms.29 Bar, 0.005 Knuc.
A whole-cell hydrolysate of strain IR73-Li102T contained meso-diaminopimelic acid, arabinose and galactose (wall chemotype IV).33 The major menaquinone was observed only a MK-8 (H4) and minor menaquinones were not observed. The predominant polar lipids were phosphatidylethanolamine, phosphatidylcholine, phosphatidylinositol and diphosphatidylglycerol (phospholipid type PIII).34 The cellular fatty acid profiles of strain IR73-Li102T and its three closest phylogenetic relatives are summarized in Table 1. In contrast to A. chiangmaiensis, A. corticola and A. rishiriensis, the only major fatty acid (>10% of the total) detected in strain IR73-Li102T was iso-C16:0 (58.1%), which represents a differentiating characteristic of these related species. The DNA G+C content of strain IR73-Li102T was 74.0 mol%. on the basis of the phylogenetic and chemotaxonomic analyzes, strain IR73-Li102T was identified as a member of the genus Actinomycetospora. Interestingly, the previously reported Actinomycetospora species were mainly isolated from subtropical/tropical region.9, 10 A similar tendency was also reported by González et al.3 It is considered that the abundance or diversity of Actinomycetospora correlated to the climate. However, ecological futures of lichen-associated actinomycetes, including Actinomycetospora were not clarified yet. The further characterization of the strain IR73-Li102T belonging to the genus Actinomycetes was examined on the basis of the comparisons with the three phylogenetically-related species.
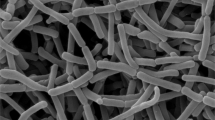
The strain IR73-Li102T formed cream–yellow aerial mycelium and bud-like spores, similar in appearance to those of A. chiangmaiensis NBRC 104400T, directly from vegetative mycelium (Figure 2). The spores were oval and rod-shaped (width, 0.4–0.5 μm; length, 0.9–1.1 μm) with a smooth surface. No diffusible pigment was detected when the strain was cultured on ISP medium 6 or 7. Growth of strain IR73-Li102T occurred in the pH range 5–8 and in the presence of 0–3% NaCl (w/v), with optimum growth observed at pH 7.0 and 0% NaCl (w/v). The temperature range for growth was 20–30 °C, with the optimum temperature being 30 °C. Strain IR73-Li102T could be differentiated from its phylogenetic relatives by acid phosphatase, utilization of D-galactose and D-mannose, fatty acid profile and growth temperature (Table 2). Strain IR73-Li102T only exhibited weakly positive antimicrobial activities against Aspergillus niger NBRC 33023T In contrast, A. corticola NBRC 103689T has both antibacterial and anti-fungal activities. The detailed physiological and biochemical properties of strain IR73-Li102T are summarized in the species description below and Table 2.
Finally, the DNA–DNA hybridization values for strain IR73-Li102T with A. chiangmaiensis NBRC 104400T, A. corticola NBRC 103689T and A. rishiriensis RI109-Li102T were below 35%, which is well below the 70% cut-off point recommended for the assignment of bacterial strains to the same genomic species.35
On the basis of the phenotypic and genotypic characteristics, strain IR73-Li102T represents a novel species within the genus Actinomycetospora, for which the name A. iriomotensis sp. nov. is proposed.
Description of Actinomycetospora iriomotensis sp. nov.
Actinomycetospora iriomotensis (i’ri.o.mo.ten'sis. N.L. fem. adj. iriomotensis, of or pertaining to Iriomote Island, Okinawa, Japan, where the organism was originally isolated).
Aerobic, Gram-positive, non-motile actinomycete, which forms oval and rod-shaped spores on aerial mycelium. Colonies are powdery surface and have cream–yellow aerial mycelium. The substrate mycelium is long, well developed and branched. Contains meso-diaminopimelic acids, and arabinose and galactose are present in whole-cell hydrolysates. The major fatty acid (>10% of total) is iso-C16:0. The polar lipid profile consists of phosphatidylethanolamine, phosphatidylcholine, phosphatidylinositol and diphosphatidylglycerol. MK-8 (H4) is the major menaquinone. Melanoid pigments are not formed on ISP medium 6 or 7. Growth occurs at 20–30 °C, but not at 15, 37 or 45 °C. Growth occurs in the presence of 0–3% NaCl (w/v) and at an initial pH of 5–8, with an optimum at pH 7. Aesculin, urea and starch are hydrolyzed, but not gelatin. Nitrate is not reduced. D-trehalose, D-turanose, D-fructose, D-glucose, glycerol, D-maltose, D-mannitol, D-sorbitol, D-sucrose and xylitol are used as the sole carbon source, but not erythritol, L-arabitol, D-arabinose, D-galactose, D-mannose or D-melezitose. In the API ZYM and Coryne systems (bioMérieux), tests for esterase (C4) and esterase lipase (C8) are positive. Naphthol-AS-BI-phosphohydrolase is weakly positive. Activities of lipase (C14), valine arylamidase, cystine arylamidase, pyrrolidonyl arylamidase, trypsin, α-chymotrypsin, acid phosphatase, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, α-fucosidase and pyrazinamidase are negative. The DNA G+C content of the type strain is 74.0 mol%.
The type strain is IR73-Li102T (=NBRC 106365T = KCTC 19782T), isolated from a lichen sample collected on Iriomote Island, Okinawa, Japan.
Accession codes
References
Mandrioli, P., Caneva, G. & Sabbioni, C. Cultural Heritage and Aerobiology: Methods and Measurement Techniques for Biodeterioration Monitoring (Springer, Verlag Berlin Heidelberg, 2003).
Nash, T. H. Lichen Biology (Cambridge University Press, Cambridge, UK, 1996).
González, I., Ayuso-Sacido, A., Anderson, A. & Genilloud, O. Actinomycetes isolated from lichens: evaluation of their diversity and detection of biosynthetic gene sequences. FEMS Microbiol. Ecol. 54, 401–415 (2005).
Cardinale, M., Puglia, A. M. & Grube, M. Molecular analysis of lichen-associated bacterial communities. FEMS Microbiol. Ecol. 57, 484–495 (2006).
Motohashi, K., Takagi, M., Yamamura, H., Hayakawa, M. & Shin-ya, K. A new angucycline and a new butenolide isolated from lichen-derived Streptomyces spp. J. Antibiot. 63, 545–548 (2010).
Li, B., Xie, C.- H. & Yokota, A. Nocardioides exalbidus sp. nov., a novel actinomycete isolated from lichen in Izu-Oshima Island, Japan. Actinomycetologica 21, 22–26 (2007).
An, S.- Y., Xiao, T. & Yokota, A. Leifsonia lichenia sp. nov., isolated from lichen in Japan. J. Gen. Appl. Microbiol. 55, 339–343 (2009).
Yamamura, H. et al. Actinomycetospora rishiriensis sp. nov., an actinomycete isolated from a lichen. Int. J. Syst. Evol. Microbiol. (e-pub ahead of print 10 December 2010; doi:10.1099/ijs.0.028753-0).
Jiang, Y. et al. Actinomycetospora chiangmaiensis gen. nov., sp. nov., a new member of the family Pseudonocardiaceae. Int. J. Syst. Evol. Microbiol. 58, 408–413 (2008).
Tamura, T. et al. Actinomycetospora chibensis sp. nov., Actinomycetospora chloros sp. nov., Actinomycetospora cinnamomea sp. nov., Actinomycetospora corticola sp. nov., Actinomycetospora lutea sp. nov., Actinomycetospora straminea sp. nov. and Actinomycetospora succinea sp. nov. and emendation of the genus Actinomycetospora. Int. J. Syst. Evol. Microbiol. (e-pub ahead of print 9 July 2010; doi:10.1099/ijs.0.018366-0).
Hayakawa, M. & Nonomura, H. Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J. Ferment. Technol. 65, 501–509 (1987).
Hayakawa, M., Iino, S. & Nonomura, H. Heavy metal resistance and melanoid pigment production in the streptomycete flora of copper-polluted vineyard soils. Hakkokogaku 60, 1–9 (1982).
Gerhardt, P. Manual of Methods for General Bacteriology (American Society for Microbiology, Washington, DC, 1981).
Shirling, E. B. & Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340 (1966).
Williams, S. T. et al. Numerical classication of Streptomyces and related taxa. J. Gen. Microbiol. 129, 1743–1813 (1983).
Gordon, R. E. & Mihm, J. M. Identification of Nocardia caviae (Erikson) nov. comb. Ann. NY. Acad. Sci. 98, 628–636 (1962).
Hasegawa, T., Takizawa, M. & Tanida, S. A rapid analysis for chemical grouping of aerobic actinomycetes. J. Gen. Appl. Microbiol. 29, 319–322 (1983).
Schaal, K. P. Chemical Methods in Bacterial Systematics (Academic Press, London, 1985).
Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids. Microbial ID, Inc. (Newark, Delaware, 1990).
Minnikin, D. E. et al. An integrated procedure for the extraction of bacterial isoprenoid quinines and polar lipids. J. Microbiol. Methods 2, 233–241 (1984).
Tamaoka, J. & Komagata, K. Determination of DNA base composition by reversed- phase high-performance liquid chromatography. FEMS Microbiol. Lett. 25, 125–128 (1984).
Saito, H. & Miura, K.- I. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta. 72, 619–629 (1963).
Hatano, K., Nishii, T. & Kasai, H. Taxonomic re-evaluation of whorl-forming Streptomyces (formerly Streptoverticillium) species by using phenotype, DNA–DNA hybridization and sequences of gyrB, and proposal of Streptomyces luteireticuli (ex Katoh and Arai 1957) corring., sp. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 53, 1519–1529 (2003).
Tamura, T. & Hatano, K. Phylogenetic analysis of the genus Actinoplanes and transfer of Actinoplanes minutisporangius Ruan et al. 1986 and ‘Actinoplanes aurantiacus’ to Cryptosporangium minutisporangium comb. nov. and Cryptosporangium aurantiacum sp. nov. Int. J. Syst. Evol. Microbiol. 51, 2119–2125 (2001).
Chun, J. et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57, 2259–2261 (2007).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Takahashi, K. & Nei, M. Efficiencies of fast algorithms of phylogenetic inference under the criteria of maximum parsimony, minimum evolution, and maximum likelihood when a large number of sequences are used. Mol. Biol. Evol. 17, 1251–1258 (2000).
Guindon, S., Lethiec, F., Duroux, P. & Gascuel, O. PHYML Online–a web server for fast maximum likelihood-based phylogenetic inference (Web Server issue). Nucleic Acids Res. 33, W557–W559 (2005).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985).
Kusunoki, S. et al. Application of calorimetric microdilution plate hybridization for rapid generic identification of 22 Mycobacterium species. J. Clin. Microbiol. 29, 1596–1603 (1991).
Lechevalier, M. P. & Lechevalier, H. A. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int. J. Syst. Evol. Microbiol. 20, 435–443 (1970).
Lechevalier, M. P., De Bivre, C. & Lechevalier, H. A. Chemotaxonomy of aerobic actinomycetes: phospholipid composition. Biochem. Syst. Ecol. 5, 249–260 (1977).
Wayne, L. G. et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464 (1987).
Acknowledgements
We are grateful to Mr Yuya Sakuraki for technical assistance. This study was supported in part by a research grant from the Institute for Fermentation, Osaka (IFO), Japan. We are grateful to Dr Jean P Euzéby for support with nomenclature.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yamamura, H., Ashizawa, H., Nakagawa, Y. et al. Actinomycetospora iriomotensis sp. nov., a novel actinomycete isolated from a lichen sample. J Antibiot 64, 289–292 (2011). https://doi.org/10.1038/ja.2011.15
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2011.15
Keywords
This article is cited by
-
Actinomycetospora callitridis sp. nov., an endophytic actinobacterium isolated from the surface-sterilised root of an Australian native pine tree
Antonie van Leeuwenhoek (2019)
-
Diversity, Antimicrobial Activity, and Biosynthetic Potential of Cultivable Actinomycetes Associated with Lichen Symbiosis
Microbial Ecology (2017)
-
Lichens as natural sources of biotechnologically relevant bacteria
Applied Microbiology and Biotechnology (2016)
-
Nutrient scavenging activity and antagonistic factors of non-photobiont lichen-associated bacteria: a review
World Journal of Microbiology and Biotechnology (2016)
-
Actinomycetospora atypica sp. nov., a novel soil actinomycete and emended description of the genus Actinomycetospora
Antonie van Leeuwenhoek (2014)