Abstract
In Japan, breast cancer is the most common cancer among women and the second leading cause of cancer death among women worldwide. To identify genetic variants associated with the disease susceptibility, we performed a genome-wide association study (GWAS) using a total of 1086 Japanese female patients with hormonal receptor-positive (HRP) breast cancer and 1816 female controls. We selected 33 single-nucleotide polymorphisms (SNPs) with suggestive associations in GWAS (P-value of <1 × 10−4) as well as 4 SNPs that were previously implicated their association with breast cancer for further replication by an independent set of 1653 cases and 2797 controls. We identified significant association of the disease with a SNP rs6788895 (Pcombined of 9.43 × 10−8 with odds ratio (OR) of 1.22) in the SIAH2 (intron of seven in absentia homolog 2) gene on chromosome 3q25.1 where the involvement in estrogen-dependent diseases was suggested. In addition, rs3750817 in intron 2 of the fibroblast growth factor receptor 2 gene, which was reported to be associated with breast cancer susceptibility, was significantly replicated with Pcombined of 8.47 × 10−8 with OR=1.22. Our results suggest a novel susceptibility locus on chromosome 3q25.1 for a HRP breast cancer.
Similar content being viewed by others
Introduction
Nearly 70% of breast cancer is known to be hormone dependent, as estrogen and progesterone have key roles both in the development and progression of the disease.1, 2 The exposures to higher level and/or for longer period of estrogen such as early menarche, late menopause, late age at first pregnancy, nulliparity, postmenopausal obesity and high serum estrogen level in postmenopausal women is considered to be risk factors for breast cancer.3, 4, 5 Furthermore, progestin, synthetic progesterone, was shown to markedly increase the risk of breast cancer in postmenopausal women when this hormonal therapy was provided for >10 years.6 In Japan, breast cancer is the most common cancer among women and its incidence has been doubled in both pre- and postmenopausal women in the last 20 years, mainly as an estrogen receptor-positive subgroup.7 Although hormone therapy and radiotherapy are effective, cancer cells often become resistant to these treatments; nearly half of estrogen receptor-positive breast cancer patients at an advanced stage suffer from recurrence8, 9, 10 and only one-third of hormonal receptor-positive (HRP) patients with metastatic disease respond to radiotherapy.11 Therefore, new therapeutic options for the disease are eagerly awaited.
The aim of this study is to identify the genetic factors susceptible to HRP breast cancer in the Japanese population and should facilitate the development of novel approaches to prevent and/or treat breast cancer.
Materials and methods
Samples
Characteristics of study subjects are shown in Table 1. Most of the breast cancer cases and all the controls in this study were registered in the BioBank Japan, which begun in 2003 with the goal of collecting DNA and serum samples, along with clinical information from 300 000 individuals who were diagnosed to have any of 47 different diseases from a collaborative network of 66 hospitals in Japan. All cases were diagnosed to have a HRP breast cancer by the following examinations: examination of breast tissue (biopsy or cytology), estrogen receptor and progesterone receptor positivities were evaluated by immunohistochemistry. For the genome-wide association study (GWAS) study, 1086 subjects with HRP breast cancer had been selected as cases (Table 1); 846 samples were collected from the BioBank Japan and the remaining 240 samples were collected from collaborative hospitals. Controls for the GWAS consisted of 1816 females including 231 healthy volunteers from the Midosuji Rotary Club, Osaka, Japan. In addition, we also used genome-wide screening data of 1585 female samples for 8 diseases registered in the BioBank Japan (Table 1). In the replication stage, 1547 cases were obtained from BioBank Japan and 105 cases from the collaborative hospitals. In all, 2797 female controls were registered in BioBank Japan and were genotyped in GWAS for other diseases (Table 1).
For re-sequencing analysis, we selected 2266 cases with HRP breast cancer from the BioBank Japan. We used 497 female controls with 4 diseases (hepatitis B, keloid, drug eruption and pulmonary tuberculosis) from the BioBank Japan as well as 231 healthy volunteers from the Midosuji Rotary Club, Osaka, Japan. All participating subjects provided written informed consent to participate in the study in accordance with the process approved by Ethical Committee at each of the Institute of Medical Science of the University of Tokyo and the Center for Genomic Medicine of RIKEN.
SNP genotyping
For the first stage, we genotyped 1086 female individuals with HRP breast cancer and 1816 female controls using the Illumina HumanHap 610 Genotyping BeadChip (Illumina, San Diego, CA, USA). We applied our single-nucleotide polymorphism (SNP) quality control standard (call rate of ⩾0.99 in both cases and controls, and Hardy–Weinberg equilibrium test of P<1.0 × 10−6 in controls). A total of 453 627 SNPs on autosomal chromosomes and 10 525 SNPs on X chromosome passed the quality control filters and were further analyzed. All control samples for the replication stage were genotyped using the Illumina HumanHap 610 BeadChip (female samples of three diseases as controls). All cluster plots were checked by visual inspection by trained personnel, and SNPs with ambiguous calls were excluded. For cases in the replication study, we used the multiplex PCR-based Invader assay (Third Wave Technologies).12 In addition, 22 variations resulted from re-sequencing analysis were selected and genotyped in 2266 cases and 728 female controls also using the multiplex PCR-based Invader assay (Third Wave Technologies, Madison, WI, USA).
Statistical analysis
Associations of SNPs were tested by employing the Cochran–Armitage trend test in both the GWA and replication stages. For the combined study, the simple combined method was applied. In the replication analyses, significance level was applied to be P-value of <1.35 × 10−3 (calculated as 0.05/37) by Bonferroni correction. Odds ratios (ORs) and confidence intervals were calculated using the non-susceptible allele as a reference. Heterogeneity between the GWAS and replication sets was examined using the Breslow–Day test. The genomic inflation factor (λGC) was calculated from the median of the Cochran–Armitage trend test statistics. The quantile–quantile plot of the logarithms of the genome-wide P-values was generated by the 'snpMatrix' package in R program v2.10.0 (see URLs), and the Manhattan plot was generated using Haploview v4.1 (see URLs). Haplotype analysis was performed by the use of Haploview v4.1 by considering genotyped SNPs located within 500 kb upstream or downstream of the marker SNP. In silico prediction of functional consequences of SNP was done by the use of the SNP info web server (see URLs). (Haploview software was used to analyze linkage disequilibrium (LD) values, visualize haplotype.)
Imputation
Imputation was performed by referring to the genotype data of Japanese (JPT) individuals as deposited in the Phase II HapMap database using MACH v1.0 (see URLs). Genotypes of SNPs that are located in the genomic region within 500 kb upstream or downstream of the marker SNP (the SNP that showed the strongest association with HRP breast cancer) were imputed. In the process of imputation, 50 Markov chain iterations were implemented. Imputed SNPs with an imputation quality score of r2<0.3 were excluded from the subsequent analysis.
Re-sequencing analysis
Initially, we carried out SNP discovery by using DNA samples of 96 cases with HRP breast cancer. We designed 98 sets of primers (Supplementary Table 1) using the genomic sequence information from UCSC Genome Bioinformatics data base (NM_005067) to amplify the 22 353 bps (two exons, one intron, 5′-UTR and 3′-UTR) of the genomic region corresponding to the SIAH2 (intron of seven in absentia homolog 2) gene. For each of the 96 DNA samples, PCRs were performed by using GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA, USA). We performed direct sequencing of the PCR products with the 96-capillary 3730 × l DNA Analyzer (Applied Biosystems) with Big Dye Terminators (Applied Biosystems) according to standard protocols. All amplified fragments were sequenced by two pairs of sequencing primers. Then SNPs were detected by Sequecher software v4.8 (Gene Codes, Ann Arbor, MI, USA).
Results
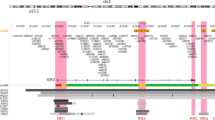
To identify genetic variants susceptible to HRP breast cancer in the Japanese population, we performed a GWAS using 1086 female patients and 1816 female controls with Illumina HumanHap 610k BeadChip (Table 1). After the quality check of SNP genotyping data, a total of 453 627 SNPs were selected for further analysis. Principal component analysis revealed that all the subjects participating in this study were clustered in the Hapmap Asian population (Supplementary Figure 1S). A quantile–quantile plot for this GWAS is shown in Supplementary Figure 2S. The genomic inflation factor (λGC) of the test statistic in this study was 1.053 indicating a very low possibility of false-positive associations resulted from the population stratification. Although no SNP achieved genome-wide significance level, 46 SNPs in various chromosomes showed suggestive association (P-values<1 × 10−4) as illustrated in Figure 1.
Among these 46 SNPs, we excluded SNPs possessing strong LD (r2>0.8) and selected 33 SNPs for replication analysis as well as 4 additional SNPs that were previously reported their association with breast cancer and showed P-value of <1.0 × 10−2 in GWAS analysis, using an independent set of 1653 female patients and 2797 female controls. Among 37 SNPs analyzed in the replication study, an SNP rs6788895 was successfully replicated with the P-value of <1.35 × 10−3 even after the Bonferroni correction (0.05/37) as shown in Table 2 and Supplementary Table 2S. Combined analysis of the results of the GWAS and the replication study suggested strong association of the locus of the SIAH2 gene on chromosome 3q25.1 (rs6788895, Pcombined of 9.43 × 10−8 with OR of 1.22, 95% confidence interval 1.13–1.31) without any significant heterogeneity between the two studies (Pheterogeneity= 2.33 × 10−01).
The SNP rs6788895 was further examined its association with the subgroups of breast cancer, an invasive papilloductal breast cancer group and a HER2-negative breast cancer group, and found significant associations with them (Pcombined=3.61 × 10−07, 6.78 × 10−06, OR=1.23, 1.21, respectively) although they did not reach to the genome-wide significant level (Supplementary Table 3S). Imputation analysis of this locus identified nine additional SNPs in strong LD (r2 of >0.8) that showed similar levels of association with rs6788895 (Figure 2a). The subsequent logistic regression analysis revealed no significant association of these nine SNPs when we accounted the effect of SNP rs6788895. The haplotype analysis found no haplotype revealing stronger association than the single SNP (Supplementary Table 4S). Although in silico prediction of the functional effect of rs6788895 identified no possible biological effect, one SNP rs2018246 showing strong LD with rs6788895 (r2=0.94), which was located about 0.7 kb upstream from the transcription initiation site of SIAH2, was indicated to be present within the binding site of multiple transcription factors such as STAT1, LEF1, PAX2, which were reported to have some implication to breast cancer.13, 14, 15, 16 The re-sequencing of 22 353 bps corresponding to the SIAH2 gene identified 10 novel genetic variations in addition to 37 genetic variations reported previously. We further genotyped 22 of the 47 variations after the exclusion of SNPs showing strong LD with the marker SNP (r2 of >0.8). As a result, we identified no genetic variant showing significant association in HRP breast cancer (Supplementary Table 5S and Supplementary Table 6S)
(a) Regional association plots of the locus associated with hormonal receptor-positive breast cancer on chromosomes 3q25.1 (intron of seven in absentia homolog 2 (SIAH2)). (b) Regional association plots of the locus associated with hormonal receptor-positive breast cancer on chromosomes 10q26 (fibroblast growth factor receptor 2 (FGFR2)). For each plot, −log10P of the Cochran–Armitage trend test of single-nucleotide polymorphisms (SNPs) in the genome-wide association study (GWAS) was plotted against relative chromosomal locations. The square and rounded signs represent imputed and genotyped SNPs, respectively. All SNPs are color coded as red (r2=0.8–1.0), orange (r2=0.6–0.8), green (r2=0.4–0.6), light blue (r2=0.4–0.6), and dark blue (r2<0.2) according to their pair wise r2 to the marker SNP. The marker SNP is represented in purple color. SNP positions followed NCBI build 36 coordinates. Estimated recombination rates (cM/Mb) are plotted as a blue line.
Furthermore, we examined the association of 37 previously reported SNPs with the HRP breast cancer17, 18, 19, 20, 21, 22, 23, 24, 25, 26 using our sample sets (Supplementary Table 7S) and found very moderate association of four genetic variants, rs1292011, rs3803662, rs2981579 and rs3750817, with HRP breast cancer in the GWAS phase (PGWAS= 5.89 × 10−02, 6.95 × 10−03, 8.68 × 10−04 and 5.03 × 10−04, respectively). Further analysis of these four SNPs identified significant replication of two SNPs, rs3750817 (Preplication=5.39 × 10−5, OR =1.22) and rs2981579 (Preplication=1.21 × 10−3, OR=1.20). Both SNPs are located within intron 2 of the fibroblast growth factor receptor 2 (FGFR2) genes. The combined analysis of the GWAS and replication phases of rs3750817 revealed strong association with Pcombined=8.47 × 10−08 (OR=1.22) and that of rs2981579 was 1.77 × 10−06 (OR=1.20) (Table 3). Imputation analysis of this locus identified three additional SNPs, rs9420318, rs11199914 and rs10736303 that showed similar levels of association with rs3750817 (Figure 2b).
Discussion
We reported here GWA and replication studies using a total of 2730 female breast cancer cases and 4613 female controls in the Japanese population to identify common genetic variants susceptible to the HRP breast cancer. The SNP rs6788895 located in the intronic region of the SIAH2 gene on chromosome 3q25.1 revealed a significant association with the HRP breast cancer (Pcombined of 9.43 × 10−08 with OR of 1.22, 95% confidence interval of 1.13–1.31). We further examined the association of rs6788895 with the subgroups of breast cancer. The analysis of two histological subgroups, an invasive papilloductal breast cancer group and a HER2-negative breast cancer group, indicated suggestive associations with Pcombined of 3.61 × 10−07 (OR=1.24) and with Pcombined of 6.78 × 10−06 (OR =1.21), respectively (Supplementary Table 3S). However, rs6788895 showed no association in the GWAS with the hormonal receptor-negative group (Ptrend of 1.03 × 10−01) or with the HER2-positive breast cancer group (Ptrend of 1.15 × 10−01).
For further characterization of the chromosome 3q25.1 locus, we imputed genotypes of SNPs that were not genotyped in the GWAS and then examined their associations with HRP breast cancer, but found no SNP showing stronger association than the marker SNP rs6788895 although several SNPs having strong LD with rs6788895 (r2>0.8) showed similar levels of associations (Figure 2a). Previous reports implicated possible roles of SIAH2 in breast carcinogenesis and described that SIAH2 expression was highly associated with estrogen receptor levels.9, 27, 28, 29 In addition, SIAH2 protein was indicated to have an essential role in the hypoxic response by regulating the hypoxia-inducible factor-α.30
Moreover, SIAH2 was known to induce ubiquitin-mediated degradation of many substrates, including proteins involved in transcriptional regulation (POU2AF1, PML and NCOR1), a cell surface receptor (DCC) and an anti-apoptotic protein (BAG1). These proteins were reported to have some relations to breast cancer by different mechanisms.31, 32, 33, 34, 35 Recent genetic studies showed that the chromosome 3q25.1 region might have a critical role in some estrogen-dependent diseases such as development of peritoneal leioyomatosis.36, 37
We also examined the association of previously reported loci with the breast cancer17, 18, 19, 20, 21, 22, 23, 24, 25, 26 using our sample sets and found very moderate association of four genetic variants in our GWAS. Further analysis of these four SNPs identified significant replication of two SNPs, rs3750817 and rs2981579 (Pcombined=8.47 × 10−8 and 1.77 × 10−06 with OR=1.22 and OR=1.20, respectively). A T allele for rs3750817 is a protective allele for both Japanese and American populations with comparable ORs (Table 3).
For characterization of the chromosome 10q26 locus, we imputed genotypes of SNPs that were not genotyped in the GWAS, and examined the associations of these SNPs with HRP breast cancer. As a result, three additional SNPs, rs9420318, rs11199914 and rs10736303 were found to have similar levels of association with rs3750817 (Figure 2b). The most strongly associated SNPs are located in intron 2 of the FGFR gene. The intron 2 region contains a highly conserved region and possess the transcription factor binding sites possibly related to the estrogen receptor signaling pathway.38 FGFR2 encodes a receptor tyrosine kinase and has an important role in human mammary epithelial-cell transformation,39, 40 suggesting that FGFR2 is a good candidate for breast cancer susceptibility. Subsequent functional analyses are thus essential to pinpoint the causal variants and genes associated with HRP breast cancer. In addition, because breast cancer is multi factorial disease, we could not exclude the possibility that some subjects with undiagnosed early stage of cancers or undiagnosed hormonal-dependent diseases or subject have diseases related to breast cancer might have been included as controls. Hence, this study might not have sufficient power to detect SNPs having very modest effects on susceptibility to HRP breast cancer. In conclusion, our findings, the verification of the association of the FGFR2 to the risk of breast cancer in the Japanese population and the novel identification of significant association of genetic variations in the SIAH2 gene, should contribute to the better understanding of the susceptibility to HRP breast cancer.
URLs
The Leading Project for Personalized Medicine, http://biobankjp.org/;
EIGENSTRATsoftwarev2.0, http://genepath.med.harvard.edu/∼reich/Software.htm;
R project v2.10.0, http://www.r-project.org/;
Haploview v4.1, http://www.broadinstitute.org/haploview/haploview;
MACH v1.0, http://www.sph.umich.edu/csg/yli/mach/index.html;
PLINK statistical software v1.06, http://pngu.mgh.harvard.edu/∼purcell/plink/;
SNP info web server, http://manticore.niehs.nih.gov/index.html.
References
Massarweh, S., Schiff, R. Resistance to endocrine therapy in breast cancer: exploiting estrogen receptor/growth factor signaling crosstalk. Endocr. Relat. Cancer 13, S15–S24 (2006).
Xu, W. -H., Shu, X. O., Long, J., Lu, W., Cai, Q., Zheng, Y. et al. Relation of FGFR2 genetic polymorphisms to the association between oral contraceptive use and the risk of breast cancer in Chinese women. Am. J. Epidemiol. 173, 923–931 (2011).
Hu, Y.-H., Nagata, C., Shimizu, H., Kaneda, N., Kashiki, Y. Association of body mass index, physical activity, and reproductive histories with breast cancer: a case-control study in Gifu, Japan. Breast Cancer Res. Treat. 43, 65–72 (1997).
Tung, H. T., Tsukuma, H., Tanaka, H., Kinoshita, N., Koyama, Y., Ajiki, W. et al. Risk factors for breast cancer in Japan, with special attention to anthropometric measurements and reproductive history. Jpn. J. Clin. Oncol. 29, 137–146 (1999).
Miyoshi, Y., Tanji, Y., Taguchi, T., Tamaki, Y., Noguchi, S. Association of serum estrone levels with estrogen receptor-positive breast cancer risk in postmenopausal Japanese women. Clin. Cancer Res. 9, 2229–2233 (2003).
Colditz, G. A., Rosner, B. Cumulative risk of breast cancer to age 70 years according to risk factor status: data from the Nurses' Health Study. Am. J. Epidemiol. 152, 950–964 (2000).
Yamashita, H., Iwase, H., Toyama, T., Takahashi, S., Sugiura, H., Yoshimoto, N. et al. Estrogen receptor-positive breast cancer in Japanese women: trends in incidence, characteristics, and prognosis. Ann. Oncol. 22, 1318–1325 (2011).
Ao, A., Morrison, B. J., Wang, H., López, J. A., Reynolds, B. A., Lu, J. Response of estrogen receptor-positive breast cancer tumorspheres to antiestrogen treatments. PLoS ONE 6, e18810 (2011).
Jansen, M., Ruigrok-Ritstier, K., Dorssers, L. C., van Staveren, I. L., Look, M. P., Meijer-van Gelder, M. E. et al. Downregulation of SIAH2, an ubiquitin E3 ligase, is associated with resistance to endocrine therapy in breast cancer. Breast Cancer Res. Treat. 116, 263–271 (2009).
Kiyotani, K., Mushiroda, T., Tsunoda, T., Morizono, T., Hosono, N., Kubo, M. et al. A genome-wide association study identifies locus at 10q22 associated with clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients in Japanese. Hum. Mol. Genet. 21, 1665–1672 (2012).
Prat, A., Baselga, J. The role of hormonal therapy in the management of hormonal-receptor-positive breast cancer with co-expression of HER2. Nat. Clin. Prac. Oncol. 5, 531–542 (2008).
Ohnishi, Y., Tanaka, T., Ozaki, K., Yamada, R., Suzuki, H., Nakamura, Y. A high-throughput SNP typing system for genome-wide association studies. J. Hum. Genet. 46, 471–477 (2001).
Chan, S., Vermi, W., Luo, J., Lucini, L., Rickert, C., Fowler, A. et al. STAT1-deficient mice spontaneously develop estrogen receptor alpha-positive luminal mammary carcinomas. Breast Cancer. Res. 14, R16 (2012).
Holmes, K. A., Song, J. S., Liu, X. S., Brown, M., Carroll, J. S. Nkx3-1 and LEF-1 function as transcriptional inhibitors of estrogen receptor activity. Cancer Res. 68, 7380–7385 (2008).
Huang, F. -I., Chen, Y. -L., Chang, C. -L., Yuan, R. -H., Jeng, Y. -M. Hepatocyte growth factor activates Wnt pathway by transcriptional activation of LEF1 to facilitate tumor invasion. Carcinogenesis 33, 1142–1148 (2012).
Beauchemin, D., Lacombe, C., Van Themsche, C. PAX2 is activated by estradiol in breast cancer cells of the luminal subgroup selectively, to confer a low invasive phenotype. Mol. Cancer 10, 148 (2011).
Walsh, T., Casadei, S., Coats, K. H., Swisher, E., Stray, S. M., Higgins, J. et al. Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA 295, 1379–1388 (2006).
Sehrawat, B., Sridharan, M., Ghosh, S., Robson, P., Cass, C. E., Mackey, J. R. et al. Potential novel candidate polymorphisms identified in genome-wide association study for breast cancer susceptibility. Hum. Genet. 130, 529–537 (2011).
Higginbotham, K. S. P., Breyer, J. P., Bradley, K. M., Schuyler, P. A., Plummer, W. D., Freudenthal, M. E. et al. A multistage association study identifies a breast cancer genetic locus at NCOA7. Cancer Res. 71, 3881–3888 (2011).
Prentice, R. L., Huang, Y., Hinds, D. A., Peters, U., Pettinger, M., Cox, D. R. et al. Variation in the FGFR2 gene and the effects of postmenopausal hormone therapy on invasive breast cancer. Cancer Epidem. Biomarker. Prev. 18, 3079–3085 (2009).
Ghoussaini, M., Fletcher, O., Michailidou, K., Turnbull, C., Schmidt, M. K., Dicks, E. et al. Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat. Genet. 44, 312–318 (2012).
Rebbeck, T. R., DeMichele, A., Tran, T. V., Panossian, S., Bunin, G. R., Troxel, A. B. et al. Hormone-dependent effects of FGFR2 and MAP3K1 in breast cancer susceptibility in a population-based sample of post-menopausal African-American and European-American women. Carcinogenesis 30, 269–274 (2009).
Yoshimoto, N., Nishiyama, T., Toyama, T., Takahashi, S., Shiraki, N., Sugiura, H. et al. Genetic and environmental predictors, endogenous hormones and growth factors, and risk of estrogen receptor-positive breast cancer in Japanese women. Cancer Sci. 102, 2065–2072 (2011).
Fanale, D., Amodeo, V., Corsini, L. R., Rizzo, S., Bazan, V., Russo, A. Breast cancer genome-wide association studies: there is strength in numbers. Oncogene 31, 2121–2128 (2012).
Turnbull, C., Ahmed, S., Morrison, J., Pernet, D., Renwick, A., Maranian, M. et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 42, 504–507 (2010).
Hamaguchi, M., Nishio, M., Toyama, T., Sugiura, H., Kondo, N., Fujii, Y. et al. Possible difference in frequencies of genetic polymorphisms of estrogen receptor α, estrogen metabolism and P53 genes between estrogen receptor-positive and -negative breast cancers. Jpn. J. Clin. Oncol. 38, 734–742 (2008).
Perou, C. M., Sørlie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Rees, C. A. et al. Molecular portraits of human breast tumours. Nature 406, 747–752 (2000).
van ’t Veer, L. J., Dai, H., van de Vijver, M. J., He, Y. D., Hart, A. A., Mao, M. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 (2002).
Roh, M. S., Hong, S. H., Jeong, J. S., Kwon, H. C., Kim, M. C., Cho, S. H. et al. Gene expression profiling of breast cancers with emphasis of β-catenin regulation. J. Korean Med. Sci. 19, 275–282 (2004).
Chan, P., Möller, A., Liu, M. C., Sceneay, J. E., Wong, C. S., Waddell, N. et al. The expression of the ubiquitin ligase SIAH2 (seven in absentia homolog 2) is mediated through gene copy number in breast cancer and is associated with a basal-like phenotype and p53 expression. Breast Cancer Res. 13, R19 (2011).
Koren, R., Dekel, Y., Sherman, E., Weissman, Y., Dreznik, Z., Klein, B. et al. The expression of DCC protein in female breast cancer. Breast Cancer Res. Treat. 80, 215–220 (2003).
Zhen-huan, Z., Yamashita, H., Toyama, T., Yamamoto, Y., Kawasoe, T., Ibusuki, M. et al. Nuclear corepressor 1 expression predicts response to first-line endocrine therapy for breast cancer patients on relapse. Chin. Med. J. 122, 1764–1768 (2009).
Plevova, P., Bouchal, J., Fiurásková, M., Foretová, L., Navrátilová, M., Zapletalová, J. et al. PML protein expression in hereditary and sporadic breast cancer. Neoplasma 54, 263–268 (2007).
Yang, X., Hao, Y., Ding, Z., Pater, A., Tang, S.-C. Differential expression of antiapoptotic gene BAG-1 in human breast normal and cancer cell lines and tissues. Clin. Cancer Res. 5, 1816–1822 (1999).
Baysal, B. E. A high-resolution integrated map spanning the SDHD gene at 11q23: a 1.1-Mb BAC contig, a partial transcript map and 15 new repeat polymorphisms in a tumour-suppressor region. Eur. J. Human Genet. 9, 121–129 (2001).
Ordulu, Z., Dal C., P., Chong, W. W., Choy, K. W., Lee, C., Muto, M. G. et al. Disseminated peritoneal leiomyomatosis after laparoscopic supracervical hysterectomy with characteristic molecular cytogenetic findings of uterine leiomyoma. Genes Chromosom. Cancer 49, 1152–1160 (2010).
Cha, P. -C., Takahashi, A., Hosono, N., Low, S. K., Kamatani, N., Kubo, M. et al. A genome-wide association study identifies three loci associated with susceptibility to uterine fibroids. Nat. Genet. 43, 447–450 (2011).
Carroll, J. S., Meyer, C. A., Song, J., Li, W., Geistlinger, T. R., Eeckhoute, J. et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38, 1289–1297 (2006).
Moffa, A. B., Ethier, S. P. Differential signal transduction of alternatively spliced FGFR2 variants expressed in human mammary epithelial cells. J. Cell. Physiol. 210, 720–731 (2007).
Koziczak, M., Holbro, T., Hynes, N. E. Blocking of FGFR signaling inhibits breast cancer cell proliferation through downregulation of D-type cyclins. Oncogene 23, 3501–3508 (2004).
Acknowledgements
We extend our heartfelt gratitude to all individuals who kindly participated in the current study. We also thank the BioBank Japan for providing DNA samples and clinical information and the members of the Rotary Club of Osaka-Midosuji District 2660 Rotary International in Japan for supporting this study. We extend our appreciation to Dr Siew-Kee Low for her useful discussion in the statistical analysis and Dr Osman Wael Mohammed for his insightful advices. Finally, we would like to express our gratefulness to technical staff of the Laboratory for Genotyping Development in RIKEN, Laboratory of Molecular Medicine, Human Genome Center, Institute of Medical Science, the University of Tokyo and technical staff of collaborative hospitals for their assistances.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Elgazzar, S., Zembutsu, H., Takahashi, A. et al. A genome-wide association study identifies a genetic variant in the SIAH2 locus associated with hormonal receptor-positive breast cancer in Japanese. J Hum Genet 57, 766–771 (2012). https://doi.org/10.1038/jhg.2012.108
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2012.108
Keywords
This article is cited by
-
Identification of novel breast cancer susceptibility loci in meta-analyses conducted among Asian and European descendants
Nature Communications (2020)
-
Previous GWAS hits in relation to young-onset breast cancer
Breast Cancer Research and Treatment (2017)