Abstract
Human leukocyte immunoglobulin-like receptors (LILR) are a family of 11 functional genes encoding five activating (LILRA1, 2, 4–6), five inhibitory (LILRB1–5) and one soluble (LILRA3) form. The number of LILR genes is conserved among individuals, except for LILRA3 and LILRA6, which exhibit copy-number variations. The LILR genes are rapidly evolving and showing large interspecies differences, making it difficult to analyze the functions of LILR using an animal model. LILRs are expressed on various cells such as lymphoid and myeloid cells and the expression patterns are different from gene to gene. The LILR gene expression and polymorphisms have been reported to be associated with autoimmune and infectious diseases such as rheumatoid arthritis and cytomegalovirus infection. Although human leukocyte antigen (HLA) class I is a well-characterized ligand for some LILRs, non-HLA ligands have been increasingly identified in recent years. LILRs have diverse functions, including the regulation of inflammation, immune tolerance, cell differentiation and nervous system plasticity. This review focuses on the genetic and functional diversity of the LILR family.
Similar content being viewed by others
Introduction
The leukocyte immunoglobulin-like receptor (LILR) multigene family is a family of paired receptors composed of inhibitory and activating forms, which are highly homologous in their extracellular regions and different in their intracellular regions.1 Inhibitory LILRs possess two or four immunoglobulin (Ig)-like domains and a long cytoplasmic tail with immunoreceptor tyrosine-based inhibition motifs. In contrast, activating LILRs have two or four Ig-like domains with short cytoplasmic tail and associate with FcRγ chain containing immunoreceptor tyrosine-based activation motifs. The soluble forms of LILRs without transmembrane and cytoplasmic regions are generated by alternative splicing, thereby serving as negative regulators.2 Structurally, LILRs are categorized into group 1 (LILRB1, LILRB2 and LILRA1–3) and group 2 (LILRB3–5 and LILRA4–6) members, based on the LILRB1 residues that interact with human leukocyte antigen (HLA) class I molecules.3 The LILR family appears to be primate-specific receptors in terms of sequence homology, although the murine homolog of LILR is proposed to be paired immunoglobulin-like receptors in view of its expression pattern, synteny with the leukocyte receptor complex region, sequence and function.4, 5, 6 The comparison of chimpanzee and human LILR genes showed that the LILR genes are rapidly evolving and exhibiting greater interspecies differences than the genome average.4 These differences make it difficult to elucidate the physiological function of LILR using an animal model. Although human experiments have been limited, it has been shown that LILR expressions are upregulated in peripheral blood leukocytes after endotoxin challenge in humans.7 Because there is a LILR that recognizes viral proteins and is a high interspecies difference in the LILR genes, LILRs might have co-evolved with pathogens. In addition to human experiments, genetic association studies will help to understand LILR functions in humans. Here we review the genetic and functional diversity of human LILRs.
Gene organization
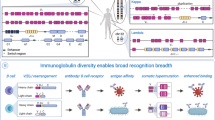
Figure 1 shows the gene organization of human LILR gene family. The LILR genes are located in the leukocyte receptor complex region on chromosome 19, which contains a number of receptors related to Ig superfamily such as killer Ig-like receptors.8 Human LILRs consist of 2 pseudogenes and 11 functional genes encoding five activating (LILRA1, 2, 4–6), five inhibitory (LILRB1–5) and one soluble (LILRA3) form. The LILR gene cluster is separated into centromeric and telomeric clusters. The centromeric and telomeric clusters are transcribed in opposite directions, from telomere to centromere and from centromere to telomere, respectively.9 The number of LILR genes is conserved among individuals, except for LILRA3 and LILRA6, which show variation in copy number. LILRA3 exhibits a presence or absence variation owing to a large deletion that removes almost the entire gene, and it is one of the genes that shows the most extreme differences in copy number among the HapMap populations.10, 11 East Asian populations have high frequency (up to 84%) of the LILRA3-deleted allele, compared with that in European (17%) and African (7%) populations.12 In addition, nonfunctional alleles containing premature stop codons have been detected in East Asians.13 These nonfunctional alleles are estimated to have been maintained for a long time in humans, suggesting that balancing selection has been acting on this locus.13 By contrast, the copy number of LILRA6 observed in an individual ranges from one to six, which can be explained by the deletion or duplication of this gene.14, 15 These LILRA6 copy-number variations contribute to the bulk of the increased African diversity.11
Gene organization and genetic associations of the LILR genes. Open and filled boxes represent the LILR genes with activating and inhibitory forms, respectively. The gene order does not reflect the actual distance. Several genes that lie between centromeric and telomeric LILR gene clusters are omitted from this figure. The gene number of LILRA6 and LILRA3 differs between haplotypes. Horizontal and vertical arrows indicate the transcriptional direction and the positions of genetic associations, respectively. CK, creatine kinase; HDL-C, high-density lipoprotein cholesterol; SS, Sjögren's syndrome; MS, multiple sclerosis; MPA, microscopic polyangiitis.
LILRB1
LILRB1 shows a broad expression pattern and is expressed on T, B, NK and myeloid cells.16, 17, 18 In contrast to homogeneous LILRB1 expression on myeloid and B cells, LILRB1 dim subsets are observed in NK cells. The frequency of LILRB1 dim NK cells varies among individuals and was significantly associated with three linked single-nucleotide polymorphisms (SNPs) in the promoter region.19 Furthermore, the frequency of LILRB1-positive NK cells in an individual fluctuates over time.20 Moreover, the cell surface expression intensity differs among LILRB1 haplotypes.21 A low-expressing LILRB1 haplotype is significantly associated with rheumatoid arthritis (RA) susceptibility in HLA-DRB1 shared epitope negative subjects, possibly because of insufficient inhibitory signaling in leukocytes.21 HLA class I is the most characterized ligand of LILRB1.16, 18, 22, 23, 24 The binding of LILRB1 to HLA class I is dependent on the nonpolymorphic α3 domain of HLA class I heavy chain and β2 microglobulin (β2 m).3, 25 Thus, LILRB1 recognizes a broad range of HLA class I alleles; however, does not recognize the β2 m-free forms of HLA class I.26 Furthermore, LILRB1 has been shown to compete with CD8 for HLA class I binding, suggesting that LILRB1 modulates CD8-positive T-cells by blocking the CD8 binding.25 HLA class I molecules are important ligands for NK cells to avoid killing self cells that expresses self HLA class I but to eliminate nonself or missing-self cells that does not express self HLA class I.27 However, this missing-self response by LILRB1 in LILRB1-single-positive NK cells that do not express other receptors for HLA class I is very low, probably because of the dependency of LILRB1 signaling on coexpressed killer Ig-like receptors.28, 29 In addition, LILRB1 recognizes the human cytomegalovirus (HCMV) HLA class I homolog UL18.17 This interaction was initially thought to be one of the immune evasion strategies by HCMV. However, the function of UL18 has remained controversial because UL18 inhibited LILRB1-positive cells but activated LILRB1-negative NK cells as well as LILRB1- and CD8-positive T-cells.30, 31 Although the involvement of UL18 is unknown, LILRB1 upregulation is correlated with the reactivation of HCMV after lung transplantation, suggesting that LILRB1 expression is a useful marker for HCMV disease.32 Besides HCMV, dengue virus also exploits LILRB1 to escape an antiviral response for enhanced replication.33 However, the LILRB1 ligand on dengue virus remains unidentified.
LILRB2
LILRB2 is expressed on myeloid cells and hematopoietic stem cells, but not T, B or NK cells.22, 34, 35 The murine homolog of LILRB2 appears to be paired immunoglobulin-like receptors-B, the only inhibitory form of the murine paired immunoglobulin-like receptors family with respect to expression pattern, ligands and function. Therefore, paired immunoglobulin-like receptors-B-deficient mice are often used for the analysis of LILRB2 function.34, 36, 37 LILRB2 SNPs are markedly differentiated between East Asians, including Japanese and Chinese, and non-East Asians, including Europeans and Africans.12 The LILRB2 alleles commonly found in East Asians are significantly associated with low levels of cell surface expression in monocytes. Moreover, LILRB2 alleles are in linkage disequilibrium with LILRA3 deletion polymorphism, and the region shows evidence of positive selection.12 Similar to LILRB1, LILRB2 recognizes a broad range of HLA class I alleles as ligands.18, 22, 24, 35 However, in contrast to LILRB1, LILRB2 recognizes not only β2 m-free forms of HLA class I but also heavy chain dimers including HLA-B27 and HLA-G, more strongly than classical HLA class I.26, 38 The binding of LILRB2 to HLA class I is affected by peptide antigens, and strong binding between them leads to the functional inhibition of myelomonocytic cells.39 The strength of LILRB2 binding to HLA class I alleles is correlated with odds ratios for corresponding HLA alleles determined by comparing HIV-1 controllers with noncontrollers.40 Moreover, LILRB2 recognizes native CD1d on the cell surface and in the endosomal and lysosomal compartments to block the loading of lipid antigen onto CD1d.41 In addition to these HLA or HLA-like molecules, LILRB2 recognizes non-HLA ligands, suggesting that LILRB2 has crucial roles in diverse biological functions. The binding of angiopoietin-like proteins to LILRB2 expressed on hematopoietic stem cells inhibits differentiation and supports the expansion of hematopoietic stem cells.34, 42 LILRB2 is also a receptor for the oligomeric forms of β-amyloid, which act as key mediators to trigger Alzheimer’s disease.37 In addition, myelin inhibitors including Nogo66, MAG and OMgp associate with LILRB2 and regulate axon regeneration.36
LILRB3
LILRB3 is expressed mainly on monocytes, dendritic cells (DCs) and granulocytes.43, 44, 45 The co-ligation of LILRB3 with LILRA2 or with the high-affinity receptor for IgE on basophils inhibits the release of histamine, cysteinyl leukotrienes and IL-4.44 LILRB3 is highly polymorphic in comparison with LILRB1 and LILRB2.16 A genome-wide association study identified a SNP, rs11666543, in the LILRB3 region as a genetic susceptibility locus for Takayasu’s arteritis in Turkish and North American cohorts.46 The risk allele is associated with significant reduction of LILRB3 expression, suggesting that inadequate inhibitory signaling results in uncontrolled immune response.46 Furthermore, allogeneic antibodies against LILRB3 were detected in 5.4% of hematopoietic stem cell transplant recipients, and they may be involved in the graft-versus-leukemia effect against LILRB3-expressing leukemic cells.45 Because LILRB3 is highly polymorphic, a large difference between the amino acid sequences of LILRB3 in donors and recipients might lead to alloantibody production.45
LILRB4
LILRB4 is expressed on monocytes, macrophages, DCs and plasmablast cells.47, 48 The upregulation of LILRB4 along with LILRB2 is associated with the tolerization of antigen-presenting cells.49 The tolerogenic function of LILRB4 has been extensively reviewed.50 Although a LILRB4 ligand is expressed on activated CD4-positive T-cells after allostimulation, the ligand has not been identified.51 LILRB4 is also highly polymorphic, and a nonsynonymous SNP rs11540761 in the putative signal sequence of LILRB4 is associated with cell surface expression on circulating monocytoid DCs in European-derived and Hispanic-American systemic lupus erythematosus (SLE) patients.52 The low-expression allele and cytoplasmically located SNP G allele (rs1048801) are independently associated with an increased serum type I interferon activity in SLE patients, suggesting an inhibitory role of LILRB4 in SLE.52
LILRB5
LILRB5 is expressed on NK cells, monocytes and mast cell granules.18, 53 LILRB5 has remained poorly characterized. However, a recent genome-wide association study identified a SNP, rs2361797, adjacent to LILRB5, as a genetic determinant of serum levels of creatine kinase in statin users and nonusers, suggesting a potential role of LILRB5 in the clearance of creatine kinase via the mononuclear phagocytic system in the liver.54
LILRA1
LILRA1 is expressed on monocytes and B cells.18 LILRA1 has been reported to bind preferentially to HLA-C free heavy chain but with lower affinity than that of LILRB1 and LILRB2.55 Furthermore, LILRA1 also binds to HLA-B27, which is strongly associated with spondyloarthropathies.26 However, the functional significance of these bindings has remained unclear.
LILRA2
LILRA2 is expressed on minor subsets of T- and NK cells, monocytes, macrophages, DCs and granulocytes.56 LILRA2 expression has been found to be elevated in the lesions of lepromatous patients, suggesting the pathological role of LILRA2 in leprosy.57 Moreover, the mean cell numbers of LILRA2-expressing cells in synovial tissue are correlated with the severity of RA.58 The cross-linking of LILRA2 with antibody induces cytotoxic granule proteins in eosinophils and proinflammatory cytokines in monocytes while inhibiting DC differentiation.43, 56, 59 LILRA2 does not bind to HLA class I molecules although it is categorized as a group 1 LILR member.26, 60 This nonbinding is possible because of structural differences between LILRA2 and the other group 1 LILRs despite more than 80% sequence identity.61 LILRA2 has a unique functional SNP that disrupts the splice acceptor site of intron 6, resulting in the generation of an alternative splicing isoform that lacks three amino acids in the stalk region.62 Interestingly, this short isoform is significantly associated with SLE and microscopic polyangiitis.62
LILRA3
LILRA3 is the only soluble form of LILR secreted by monocytes, B cells and subsets of T-cells and is not detected on the cell surface.63 LILRA3 expression is upregulated by IL-10 and IFN-γ, and downregulated by TNF-α.63 LILRA3 concentrations in serum from RA patients are correlated with disease activity.63 Similar to LILRA1, LILRA3 binds to HLA class I molecules with reduced affinities compared with LILRB1 and LILRB2.64 It is speculated that LILRA3 functions as an antagonist or a negative regulator of the other LILRs. LILRA3 is deficient in more than half of East Asians, indicating that LILRA3 is not essential for survival.12 However, LILRA3 deletion polymorphism has been reported to be associated with several autoimmune diseases. First, LILRA3 deficiency is a risk factor for multiple sclerosis in the German and Spanish populations but not in the Polish population.65, 66, 67 Second, homozygous LILRA3 deletion is associated with Sjögren’s syndrome in German, whereas homozygous functional LILRA3 is associated with primary Sjögren's syndrome in Chinese.68, 69 Moreover, homozygosity for the LILRA3 non-deleted allele confers susceptibility to RA and SLE.69, 70 In addition to autoimmune diseases, a genome-wide association study in Chinese men has identified a SNP, rs103294, adjacent to LILRA3, as a risk locus for prostate cancer.71 This SNP is in strong linkage disequilibrium with the LILRA3 deletion polymorphism.70 Unexpectedly, a SNP, rs386000, located between LILRB2 and LILRA3 is associated with plasma levels of high-density lipoprotein cholesterol, as revealed by a genome-wide association study.72
LILRA4
LILRA4 is selectively expressed on plasmacytoid DCs, but not on monocytes, T, B or NK cells.73 LILRA4 recognizes BST2 as a ligand and suppresses plasmacytoid DC activation, although LILRA4 is structurally an activating receptor.74 The suppressive function of LILRA4 may be explained by a concept termed inhibitory immunoreceptor tyrosine-based activation motif, the function of which may be partially explained by ligand-binding affinity.75, 76 BST2 is known to inhibit retrovirus release from infected cells and is induced in various cells by type I interferon.77 Therefore, it has been speculated that LILRA4 suppresses plasmacytoid DC by recognizing BST2 induced by type I interferon from neighboring cells in a negative-feedback manner. However, another group disputes this model.78
LILRA5
LILRA5 is expressed on monocytes and neutrophils.79 LILRA5 expression is upregulated by IL-10 and IFN-γ, but downregulated by TNF-α.80 Similar to LILRA2, the numbers of LILRA5-expressing cells in synovial tissue are correlated with disease activity score in patients with RA.80 The cross-linking of LILRA5 on monocytes induces proinflammatory cytokines.79
LILRA6
LILRA6 expression has been confirmed at the mRNA level in monocytes, but not in T, B or NK cells.15 The relative level of LILRA6/LILRB3 mRNA expression is correlated with copy numbers of the LILRA6 gene.15 LILRA6 is highly homologous to LILRB3, and polymorphisms in their extracellular domains show a marked excess of nonsynonymous over synonymous substitutions, reflecting a consequence of positive selection.14
Concluding remarks
LILR expression, ligand, function and genetic associations are summarized in Figure 1 and Table 1. Although the LILR family was identified more than 15 years ago, its biological and clinical significance has remained poorly understood. Particularly, 6 out of 11 LILRs are still orphan receptors, and the major ligands identified to date are HLA or HLA-like molecules. The recent identification of non-HLA ligands such as angiopoietin-like proteins and BST2 has revealed that LILRs recognize a variety of molecules in addition to HLA-related molecules. It will be interesting to identify the structural differences between LILR/HLA ligand and LILR/non-HLA ligand complexes. Moreover, genome-wide association studies offer great potential for the understanding of LILR that has not been elucidated by conventional immunological approaches. Further replication studies and fine mapping will be required to identify the true determinants of genetic associations. As described above, the LILR gene cluster exhibits a number of functional SNPs and copy-number variations. Therefore, further investigations of the LILR family will enable us to understand inter-individual differences in humans and their application to clinical practice.
References
Arase, H. & Lanier, L. L. Specific recognition of virus-infected cells by paired NK receptors. Rev. Med. Virol. 14, 83–93 (2004).
Jones, D. C., Roghanian, A., Brown, D. P., Chang, C., Allen, R. L., Trowsdale, J. et al. Alternative mRNA splicing creates transcripts encoding soluble proteins from most LILR genes. Eur. J. Immunol. 39, 3195–3206 (2009).
Willcox, B. E., Thomas, L. M. & Bjorkman, P. J. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat Immunol. 4, 913–919 (2003).
Canavez, F., Young, N. T., Guethlein, L. A., Rajalingam, R., Khakoo, S. I., Shum, B. P. et al. Comparison of chimpanzee and human leukocyte Ig-like receptor genes reveals framework and rapidly evolving genes. J. Immunol. 167, 5786–5794 (2001).
Slukvin II, Grendell, R. L., Rao, D. S., Hughes, A. L. & Golos, T. G. Cloning of rhesus monkey LILRs. Tissue Antigens. 67, 331–337 (2006).
Takai, T. Paired immunoglobulin-like receptors and their MHC class I recognition. Immunology 115, 433–440 (2005).
Talwar, S., Munson, P. J., Barb, J., Fiuza, C., Cintron, A. P., Logun, C. et al. Gene expression profiles of peripheral blood leukocytes after endotoxin challenge in humans. Physiol. Genomics. 25, 203–215 (2006).
Kelley, J., Walter, L. & Trowsdale, J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet. 1, 129–139 (2005).
Wende, H., Volz, A. & Ziegler, A. Extensive gene duplications and a large inversion characterize the human leukocyte receptor cluster. Immunogenetics 51, 703–713 (2000).
Torkar, M., Haude, A., Milne, S., Beck, S., Trowsdale, J. & Wilson, M. J. Arrangement of the ILT gene cluster: a common null allele of the ILT6 gene results from a 6.7-kbp deletion. Eur. J. Immunol. 30, 3655–3662 (2000).
Sudmant, P. H., Kitzman, J. O., Antonacci, F., Alkan, C., Malig, M., Tsalenko, A. et al. Diversity of human copy number variation and multicopy genes. Science 330, 641–646 (2010).
Hirayasu, K., Ohashi, J., Tanaka, H., Kashiwase, K., Ogawa, A., Takanashi, M. et al. Evidence for natural selection on leukocyte immunoglobulin-like receptors for HLA class I in Northeast Asians. Am. J. Hum. Genet. 82, 1075–1083 (2008).
Hirayasu, K., Ohashi, J., Kashiwase, K., Takanashi, M., Satake, M., Tokunaga, K. et al. Long-term persistence of both functional and non-functional alleles at the leukocyte immunoglobulin-like receptor A3 (LILRA3) locus suggests balancing selection. Hum. Genet. 119, 436–443 (2006).
Lopez-Alvarez, M. R., Jones, D. C., Jiang, W., Traherne, J. A. & Trowsdale, J. Copy number and nucleotide variation of the LILR family of myelomonocytic cell activating and inhibitory receptors. Immunogenetics 66, 73–83 (2014).
Bashirova, A. A., Apps, R., Vince, N., Mochalova, Y., Yu, X. G. & Carrington, M. Diversity of the human LILRB3/A6 locus encoding a myeloid inhibitory and activating receptor pair. Immunogenetics 66, 1–8 (2014).
Colonna, M., Navarro, F., Bellon, T., Llano, M., Garcia, P., Samaridis, J. et al. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J. Exp. Med. 186, 1809–1818 (1997).
Cosman, D., Fanger, N., Borges, L., Kubin, M., Chin, W., Peterson, L. et al. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity 7, 273–282 (1997).
Borges, L., Hsu, M. L., Fanger, N., Kubin, M. & Cosman, D. A family of human lymphoid and myeloid Ig-like receptors, some of which bind to MHC class I molecules. J. Immunol. 159, 5192–5196 (1997).
Davidson, C. L., Li, N. L. & Burshtyn, D. N. LILRB1 polymorphism and surface phenotypes of natural killer cells. Hum. Immunol. 71, 942–949 (2010).
Li, N. L., Davidson, C. L., Humar, A. & Burshtyn, D. N. Modulation of the inhibitory receptor leukocyte Ig-like receptor 1 on human natural killer cells. Front. Immunol 2, 46 (2011).
Kuroki, K., Tsuchiya, N., Shiroishi, M., Rasubala, L., Yamashita, Y., Matsuta, K. et al. Extensive polymorphisms of LILRB1 (ILT2, LIR1) and their association with HLA-DRB1 shared epitope negative rheumatoid arthritis. Hum. Mol. Genet 14, 2469–2480 (2005).
Fanger, N. A., Cosman, D., Peterson, L., Braddy, S. C., Maliszewski, C. R. & Borges, L. The MHC class I binding proteins LIR-1 and LIR-2 inhibit Fc receptor-mediated signaling in monocytes. Eur. J. Immunol. 28, 3423–3434 (1998).
Navarro, F., Llano, M., Bellon, T., Colonna, M., Geraghty, D. E. & Lopez-Botet, M. The ILT2(LIR1) and CD94/NKG2A NK cell receptors respectively recognize HLA-G1 and HLA-E molecules co-expressed on target cells. Eur. J. Immunol. 29, 277–283 (1999).
Lepin, E. J., Bastin, J. M., Allan, D. S., Roncador, G., Braud, V. M., Mason, D. Y. et al. Functional characterization of HLA-F and binding of HLA-F tetramers to ILT2 and ILT4 receptors. Eur. J. Immunol. 30, 3552–3561 (2000).
Shiroishi, M., Tsumoto, K., Amano, K., Shirakihara, Y., Colonna, M., Braud, V. M. et al. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc. Natl Acad. Sci. USA. 100, 8856–8861 (2003).
Allen, R. L., Raine, T., Haude, A., Trowsdale, J. & Wilson, M. J. Leukocyte receptor complex-encoded immunomodulatory receptors show differing specificity for alternative HLA-B27 structures. J. Immunol. 167, 5543–5547 (2001).
Karre, K. Natural killer cell recognition of missing self. Nat. Immunol. 9, 477–480 (2008).
Yawata, M., Yawata, N., Draghi, M., Partheniou, F., Little, A. M. & Parham, P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood 112, 2369–2380 (2008).
Kirwan, S. E. & Burshtyn, D. N. Killer cell Ig-like receptor-dependent signaling by Ig-like transcript 2 (ILT2/CD85j/LILRB1/LIR-1). J. Immunol. 175, 5006–5015 (2005).
Prod'homme, V., Griffin, C., Aicheler, R. J., Wang, E. C., McSharry, B. P., Rickards, C. R. et al. The human cytomegalovirus MHC class I homolog UL18 inhibits LIR-1+ but activates LIR-1- NK cells. J. Immunol. 178, 4473–4481 (2007).
Saverino, D., Ghiotto, F., Merlo, A., Bruno, S., Battini, L., Occhino, M. et al. Specific recognition of the viral protein UL18 by CD85j/LIR-1/ILT2 on CD8+ T cells mediates the non-MHC-restricted lysis of human cytomegalovirus-infected cells. J. Immunol. 172, 5629–5637 (2004).
Berg, L., Riise, G. C., Cosman, D., Bergstrom, T., Olofsson, S., Karre, K. et al. LIR-1 expression on lymphocytes, and cytomegalovirus disease in lung-transplant recipients. Lancet 361, 1099–1101 (2003).
Chan, K. R., Ong, E. Z., Tan, H. C., Zhang, S. L., Zhang, Q., Tang, K. F. et al. Leukocyte immunoglobulin-like receptor B1 is critical for antibody-dependent dengue. Proc. Natl. Acad. Sci. USA. 111, 2722–2727 (2014).
Zheng, J., Umikawa, M., Cui, C., Li, J., Chen, X., Zhang, C. et al. Inhibitory receptors bind ANGPTLs and blood stem cells and leukaemia development. Nature 485, 656–660 (2012).
Colonna, M., Samaridis, J., Cella, M., Angman, L., Allen, R. L., O'Callaghan, C. A. et al. Human myelomonocytic cells express an inhibitory receptor for classical and nonclassical MHC class I molecules. J. Immunol. 160, 3096–3100 (1998).
Atwal, J. K., Pinkston-Gosse, J., Syken, J., Stawicki, S., Wu, Y., Shatz, C. et al. PirB is a functional receptor for myelin inhibitors of axonal regeneration. Science 322, 967–970 (2008).
Kim, T., Vidal, G. S., Djurisic, M., William, C. M., Birnbaum, M. E., Garcia, K. C. et al. Human LilrB2 is a beta-amyloid receptor and its murine homolog PirB regulates synaptic plasticity in an Alzheimer's model. Science 341, 1399–1404 (2013).
Shiroishi, M., Kuroki, K., Rasubala, L., Tsumoto, K., Kumagai, I., Kurimoto, E. et al. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d). Proc. Natl Acad. Sci. USA. 103, 16412–16417 (2006).
Lichterfeld, M., Kavanagh, D. G., Williams, K. L., Moza, B., Mui, S. K., Miura, T. et al. A viral CTL escape mutation leading to immunoglobulin-like transcript 4-mediated functional inhibition of myelomonocytic cells. J. Exp. Med. 204, 2813–2824 (2007).
Bashirova, A. A., Martin-Gayo, E., Jones, D. C., Qi, Y., Apps, R., Gao, X. et al. LILRB2 interaction with HLA class I correlates with control of HIV-1 infection. PLoS Genet. 10, e1004196 (2014).
Li, D., Wang, L., Yu, L., Freundt, E. C., Jin, B., Screaton, G. R. et al. Ig-like transcript 4 inhibits lipid antigen presentation through direct CD1d interaction. J Immunol. 182, 1033–1040 (2009).
Deng, M., Lu, Z., Zheng, J., Wan, X., Chen, X., Hirayasu, K. et al. A motif in LILRB2 critical for Angptl2 binding and activation. Blood 124, 924–935 (2014).
Tedla, N., Bandeira-Melo, C., Tassinari, P., Sloane, D. E., Samplaski, M., Cosman, D. et al. Activation of human eosinophils through leukocyte immunoglobulin-like receptor 7. Proc. Natl Acad. Sci. USA. 100, 1174–1179 (2003).
Sloane, D. E., Tedla, N., Awoniyi, M., Macglashan, D. W. Jr., Borges, L., Austen, K. F. et al. Leukocyte immunoglobulin-like receptors: novel innate receptors for human basophil activation and inhibition. Blood 104, 2832–2839 (2004).
Pfistershammer, K., Lawitschka, A., Klauser, C., Leitner, J., Weigl, R., Heemskerk, M. H. et al. Allogeneic disparities in immunoglobulin-like transcript 5 induce potent antibody responses in hematopoietic stem cell transplant recipients. Blood 114, 2323–2332 (2009).
Renauer, P., Saruhan-Direskeneli, G., Coit, P., Adler, A., Aksu, K., Keser, G. et al. Genome-wide association study identifies susceptibility loci in IL6, RPS9/LILRB3, and an intergenic locus on chromosome 21q22 in Takayasu's arteritis. Arthritis Rheumatol 67, 1361–1368 (2015).
Cella, M., Dohring, C., Samaridis, J., Dessing, M., Brockhaus, M., Lanzavecchia, A. et al. A novel inhibitory receptor (ILT3) expressed on monocytes, macrophages, and dendritic cells involved in antigen processing. J. Exp. Med. 185, 1743–1751 (1997).
Inui, M., Hirota, S., Hirano, K., Fujii, H., Sugahara-Tobinai, A., Ishii, T. et al. Human CD43+ B cells are closely related not only to memory B cells phenotypically but also to plasma blasts developmentally in healthy individuals. Int. Immunol. (e-pub ahead of print 5 March 2015; pii: dxv009).
Chang, C. C., Ciubotariu, R., Manavalan, J. S., Yuan, J., Colovai, A. I., Piazza, F. et al. Tolerization of dendritic cells by T(S) cells: the crucial role of inhibitory receptors ILT3 and ILT4. Nat. Immunol. 3, 237–243 (2002).
Vlad, G., Chang, C. C., Colovai, A. I., Vasilescu, E. R., Cortesini, R. & Suciu-Foca, N. Membrane and soluble ILT3 are critical to the generation of T suppressor cells and induction of immunological tolerance. Int. Rev. Immunol. 29, 119–132 (2010).
Kim-Schulze, S., Scotto, L., Vlad, G., Piazza, F., Lin, H., Liu, Z. et al. Recombinant Ig-like transcript 3-Fc modulates T cell responses via induction of Th anergy and differentiation of CD8+ T suppressor cells. J. Immunol. 176, 2790–2798 (2006).
Jensen, M. A., Patterson, K. C., Kumar, A. A., Kumabe, M., Franek, B. S. & Niewold, T. B. Functional genetic polymorphisms in ILT3 are associated with decreased surface expression on dendritic cells and increased serum cytokines in lupus patients. Ann. Rheum. Dis. 72, 596–601 (2013).
Tedla, N., Lee, C. W., Borges, L., Geczy, C. L. & Arm, J. P. Differential expression of leukocyte immunoglobulin-like receptors on cord-blood-derived human mast cell progenitors and mature mast cells. J. Leukoc. Biol. 83, 334–343 (2008).
Dube, M. P., Zetler, R., Barhdadi, A., Brown, A. M., Mongrain, I., Normand, V. et al. CKM and LILRB5 are associated with serum levels of creatine kinase. Circ. Cardiovasc. Genet 7, 880–886 (2014).
Jones, D. C., Kosmoliaptsis, V., Apps, R., Lapaque, N., Smith, I., Kono, A. et al. HLA class I allelic sequence and conformation regulate leukocyte Ig-like receptor binding. J. Immunol. 186, 2990–2997 (2011).
Lu, H. K., Mitchell, A., Endoh, Y., Hampartzoumian, T., Huynh, O., Borges, L. et al. LILRA2 selectively modulates LPS-mediated cytokine production and inhibits phagocytosis by monocytes. PLoS One 7, e33478 (2012).
Bleharski, J. R., Li, H., Meinken, C., Graeber, T. G., Ochoa, M. T., Yamamura, M. et al. Use of genetic profiling in leprosy to discriminate clinical forms of the disease. Science 301, 1527–1530 (2003).
Tedla, N., An, H., Borges, L., Vollmer-Conna, U., Bryant, K., Geczy, C. et al. Expression of activating and inhibitory leukocyte immunoglobulin-like receptors in rheumatoid synovium: correlations to disease activity. Tissue Antigens. 77, 305–316 (2011).
Lee, D. J., Sieling, P. A., Ochoa, M. T., Krutzik, S. R., Guo, B., Hernandez, M. et al. LILRA2 activation inhibits dendritic cell differentiation and antigen presentation to T cells. J. Immunol. 179, 8128–8136 (2007).
Nakajima, H., Samaridis, J., Angman, L. & Colonna, M. Human myeloid cells express an activating ILT receptor (ILT1) that associates with Fc receptor gamma-chain. J. Immunol. 162, 5–8 (1999).
Chen, Y., Gao, F., Chu, F., Peng, H., Zong, L., Liu, Y. et al. Crystal structure of myeloid cell activating receptor leukocyte Ig-like receptor A2 (LILRA2/ILT1/LIR-7) domain swapped dimer: molecular basis for its non-binding to MHC complexes. J. Mol. Biol. 386, 841–853 (2009).
Mamegano, K., Kuroki, K., Miyashita, R., Kusaoi, M., Kobayashi, S., Matsuta, K. et al. Association of LILRA2 (ILT1, LIR7) splice site polymorphism with systemic lupus erythematosus and microscopic polyangiitis. Genes Immun. 9, 214–223 (2008).
An, H., Chandra, V., Piraino, B., Borges, L., Geczy, C., McNeil, H. P. et al. Soluble LILRA3, a potential natural antiinflammatory protein, is increased in patients with rheumatoid arthritis and is tightly regulated by interleukin 10, tumor necrosis factor-alpha, and interferon-gamma. J. Rheumatol. 37, 1596–1606 (2010).
Ryu, M., Chen, Y., Qi, J., Liu, J., Fan, Z., Nam, G. et al. LILRA3 binds both classical and non-classical HLA class I molecules but with reduced affinities compared to LILRB1/LILRB2: structural evidence. PLoS One 6, e19245 (2011).
Koch, S., Goedde, R., Nigmatova, V., Epplen, J. T., Muller, N., de Seze, J. et al. Association of multiple sclerosis with ILT6 deficiency. Genes Immun. 6, 445–447 (2005).
Ordonez, D., Sanchez, A. J., Martinez-Rodriguez, J. E., Cisneros, E., Ramil, E., Romo, N. et al. Multiple sclerosis associates with LILRA3 deletion in Spanish patients. Genes Immun. 10, 579–585 (2009).
Wisniewski, A., Wagner, M., Nowak, I., Bilinska, M., Pokryszko-Dragan, A., Jasek, M. et al. 6.7-kbp deletion in LILRA3 (ILT6) gene is associated with later onset of the multiple sclerosis in a Polish population. Hum. Immunol. 74, 353–357 (2013).
Kabalak, G., Dobberstein, S. B., Matthias, T., Reuter, S., The, Y. H., Dorner, T. et al. Association of immunoglobulin-like transcript 6 deficiency with Sjogren's syndrome. Arthritis Rheum. 60, 2923–2925 (2009).
Du, Y., Su, Y., He, J., Yang, Y., Shi, Y., Cui, Y. et al. Impact of the leucocyte immunoglobulin-like receptor A3 (LILRA3) on susceptibility and subphenotypes of systemic lupus erythematosus and Sjogren's syndrome. Ann. Rheum. Dis. (e-pub ahead of print 6 June 2014; doi:10.1136/annrheumdis-2013-204441).
Du, Y., Cui, Y., Liu, X., Hu, F., Yang, Y., Wu, X. et al. Contribution of functional LILRA3, but not nonfunctional LILRA3, to sex bias in susceptibility and severity of anti-citrullinated protein antibody-positive rheumatoid arthritis. Arthritis Rheumatol 66, 822–830 (2014).
Xu, J., Mo, Z., Ye, D., Wang, M., Liu, F., Jin, G. et al. Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nat. Genet. 44, 1231–1235 (2012).
Teslovich, T. M., Musunuru, K., Smith, A. V., Edmondson, A. C., Stylianou, I. M., Koseki, M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
Cao, W., Rosen, D. B., Ito, T., Bover, L., Bao, M., Watanabe, G. et al. Plasmacytoid dendritic cell-specific receptor ILT7-Fc epsilonRI gamma inhibits Toll-like receptor-induced interferon production. J. Exp. Med. 203, 1399–1405 (2006).
Cao, W., Bover, L., Cho, M., Wen, X., Hanabuchi, S., Bao, M. et al. Regulation of TLR7/9 responses in plasmacytoid dendritic cells by BST2 and ILT7 receptor interaction. J. Exp. Med. 206, 1603–1614 (2009).
Blank, U., Launay, P., Benhamou, M. & Monteiro, R. C. Inhibitory ITAMs as novel regulators of immunity. Immunol. Rev. 232, 59–71 (2009).
Hamerman, J. A., Tchao, N. K., Lowell, C. A. & Lanier, L. L. Enhanced Toll-like receptor responses in the absence of signaling adaptor DAP12. Nat. Immunol. 6, 579–586 (2005).
Neil, S. J., Zang, T. & Bieniasz, P. D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430 (2008).
Tavano, B., Galao, R. P., Graham, D. R., Neil, S. J., Aquino, V. N., Fuchs, D. et al. Ig-like transcript 7, but not bone marrow stromal cell antigen 2 (also known as HM1.24, tetherin, or CD317), modulates plasmacytoid dendritic cell function in primary human blood leukocytes. J. Immunol. 190, 2622–2630 (2013).
Borges, L., Kubin, M. & Kuhlman, T. LIR9, an immunoglobulin-superfamily-activating receptor, is expressed as a transmembrane and as a secreted molecule. Blood 101, 1484–1486 (2003).
Mitchell, A., Rentero, C., Endoh, Y., Hsu, K., Gaus, K., Geczy, C. et al. LILRA5 is expressed by synovial tissue macrophages in rheumatoid arthritis, selectively induces pro-inflammatory cytokines and IL-10 and is regulated by TNF-alpha, IL-10 and IFN-gamma. Eur. J. Immunol. 38, 3459–3473 (2008).
Acknowledgements
This work was supported by JSPS KAKENHI Grant Numbers 23390112, 25133705, 26117714 and 26870334.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Hirayasu, K., Arase, H. Functional and genetic diversity of leukocyte immunoglobulin-like receptor and implication for disease associations. J Hum Genet 60, 703–708 (2015). https://doi.org/10.1038/jhg.2015.64
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2015.64
This article is cited by
-
Activation of immune signals during organ transplantation
Signal Transduction and Targeted Therapy (2023)
-
Immunoglobulin-like receptors and the generation of innate immune memory
Immunogenetics (2022)
-
Characterization of LILRB3 and LILRA6 allelic variants in the Japanese population
Journal of Human Genetics (2021)
-
TARM1 contributes to development of arthritis by activating dendritic cells through recognition of collagens
Nature Communications (2021)
-
LILRB3 supports acute myeloid leukemia development and regulates T-cell antitumor immune responses through the TRAF2–cFLIP–NF-κB signaling axis
Nature Cancer (2021)