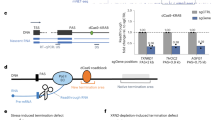
Abstract
Eukaryotic protein-encoding genes possess poly(A) signals that define the end of the messenger RNA and mediate downstream transcriptional termination by RNA polymerase II (Pol II)1. Termination could occur through an ‘anti-termination’ mechanism whereby elongation factors dissociate when the poly(A) signal is encountered, producing termination-competent Pol II2,3. An alternative ‘torpedo’ model postulated that poly(A) site cleavage provides an unprotected RNA 5′ end that is degraded by 5′ → 3′ exonuclease activities (torpedoes) and so induces dissociation of Pol II from the DNA template1,4. This model has been questioned because unprocessed transcripts read all the way to the site of transcriptional termination before upstream polyadenylation5,6,7. However, nascent transcripts located 1 kilobase downstream of the human β-globin gene poly(A) signal are associated with a co-transcriptional cleavage (CoTC) activity8 that acts with the poly(A) signal to elicit efficient transcriptional termination. The CoTC sequence is an autocatalytic RNA structure that undergoes rapid self-cleavage9. Here we show that CoTC acts as a precursor to termination by presenting a free RNA 5′ end that is recognized by the human 5′ → 3′ exonuclease Xrn2. Degradation of the downstream cleavage product by Xrn2 results in transcriptional termination, as envisaged in the torpedo model.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Proudfoot, N. J. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem. Sci. 14, 105–110 (1989)
Logan, J., Falck-Pedersen, E., Darnell, J. E. Jr & Shenk, T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc. Natl Acad. Sci. USA 84, 8306–8310 (1987)
Proudfoot, N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 16, 272–278 (2004)
Connelly, S. & Manley, J. L. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 2, 440–452 (1988)
Osheim, Y. N., Proudfoot, N. J. & Beyer, A. L. EM visualization of transcription by RNA polymerase II: downstream termination requires a poly(A) signal but not transcript cleavage. Mol. Cell 3, 379–387 (1999)
Bauren, G., Belikov, S. & Wieslander, L. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes Dev. 12, 2759–2769 (1998)
Dye, M. J. & Proudfoot, N. J. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol. Cell 3, 371–378 (1999)
Dye, M. J. & Proudfoot, N. J. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell 105, 669–681 (2001)
Teixeira, A. et al. Auto-catalytic RNA cleavage in the human β-globin pre-mRNA promotes transcription termination. Nature doi:10.1038/nature03032 (this issue)
Cerutti, H. RNA interference: travelling in the cell and gaining functions. Trends Genet. 19, 39–46 (2003)
Zhang, M. et al. Cloning and mapping of the XRN2 gene to human chromosome 20p11.1–p11.2. Genomics 59, 252–254 (1999)
Lejeune, F., Li, X. & Maquat, L. E. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12, 675–687 (2003)
Amberg, D. C., Goldstein, A. L. & Cole, C. N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 6, 1173–1189 (1992)
Henry, Y. et al. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 13, 2452–2463 (1994)
Petfalski, E., Dandekar, T., Henry, Y. & Tollervey, D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 18, 1181–1189 (1998)
Strausberg, R. L. et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl Acad. Sci. USA 99, 16899–16903 (2002)
Samarsky, D. A. et al. A small nucleolar RNA:ribozyme hybrid cleaves a nucleolar RNA target in vivo with near-perfect efficiency. Proc. Natl Acad. Sci. USA 96, 6609–6614 (1999)
Huertas, P. & Aguilera, A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12, 711–721 (2003)
Hammann, C. & Lilley, D. M. Folding and activity of the hammerhead ribozyme. Chembiochem 3, 690–700 (2002)
Stevens, A. & Poole, T. L. 5′ exonuclease 2 of Saccharomyces cerevisiae. J. Biol. Chem. 270, 16063–16069 (1995)
Torchet, C. et al. Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell 9, 1285–1296 (2002)
Kim, M. et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature doi:10.1038/nature03041 (this issue)
Adams, S. E. et al. Synthesis of a gene for the HIV transactivator protein TAT by a novel single-stranded approach involving in vivo gap repair. Nucleic Acids Res. 16, 4287–4298 (1988)
Ashe, H. L., Monks, J., Wijgerde, M., Fraser, P. & Proudfoot, N. J. Intergenic transcription and transinduction of the human β-globin locus. Genes Dev. 11, 2494–2509 (1997)
Wagner, E. J. & Garcia-Blanco, M. A. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell 10, 943–949 (2002)
Wollerton, M. C., Gooding, C., Wagner, E. J., Garcia-Blanco, M. A. & Smith, C. W. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13, 91–100 (2004)
Kadener, S., Fededa, J. P., Rosbash, M. & Kornblihtt, A. R. Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc. Natl Acad. Sci. USA 99, 8185–8190 (2002)
Acknowledgements
We thank M. Dye for expert advice on NRO analysis, M. Wollerton for advice on RNAi, I. Martianov for help with the western blot, L. Maquat and F. Lejeune for providing Xrn2 antibody, and A. Teixeira, A. Akoulitchev and members of N.J.P's laboratory for advice and encouragement. This work was supported by a Programme Grant from the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
West, S., Gromak, N. & Proudfoot, N. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432, 522–525 (2004). https://doi.org/10.1038/nature03035
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03035
This article is cited by
-
Nuclear mRNA decay: regulatory networks that control gene expression
Nature Reviews Genetics (2024)
-
CCIVR2 facilitates comprehensive identification of both overlapping and non-overlapping antisense transcripts within specified regions
Scientific Reports (2023)
-
Ceg1 depletion reveals mechanisms governing degradation of non-capped RNAs in Saccharomyces cerevisiae
Communications Biology (2023)
-
Observation of conformational changes that underlie the catalytic cycle of Xrn2
Nature Chemical Biology (2022)
-
Structural insights into nuclear transcription by eukaryotic DNA-dependent RNA polymerases
Nature Reviews Molecular Cell Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.