Abstract
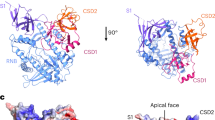
The coordinated regulation of gene expression is required for homeostasis, growth and development in all organisms. Such coordination may be partly achieved at the level of messenger RNA stability1, in which the targeted destruction of subsets of transcripts generates the potential for cross-regulating metabolic pathways. In Escherichia coli, the balance and composition of the transcript population is affected by RNase E, an essential endoribonuclease that not only turns over RNA but also processes certain key RNA precursors2,3,4,5,6,7,8,9,10. RNase E cleaves RNA internally, but its catalytic power is determined by the 5′ terminus of the substrate, even if this lies at a distance from the cutting site11,12,13,14. Here we report crystal structures of the catalytic domain of RNase E as trapped allosteric intermediates with RNA substrates. Four subunits of RNase E catalytic domain associate into an interwoven quaternary structure, explaining why the subunit organization is required for catalytic activity. The subdomain encompassing the active site is structurally congruent to a deoxyribonuclease, making an unexpected link in the evolutionary history of RNA and DNA nucleases. The structure explains how the recognition of the 5′ terminus of the substrate may trigger catalysis and also sheds light on the question of how RNase E might selectively process, rather than destroy, specific RNA precursors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Grunberg-Manago, M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33, 193–227 (1999)
Kuwano, M. et al. Gene affecting longevity of messenger RNA: a mutant of Escherichia coli with altered mRNA stability. Mol. Gen. Genet. 154, 279–285 (1977)
Ono, M. & Kuwano, M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical half life of mRNA. J. Mol. Biol. 129, 343–357 (1979)
Bernstein, J. A., Lin, P.-H., Cohen, S. N. & Lin-Chao, S. Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc. Natl Acad. Sci. USA 101, 2758–2763 (2004)
Mudd, E. A. & Higgins, C. F. Escherichia coli endoribonuclease RNase E: autoregulation of expression and site-specific cleavage of mRNA. Mol. Microbiol. 9, 557–568 (1993)
Apirion, D. & Lassar, A. B. A conditional lethal mutant of Escherichia coli which affects the processing of ribosomal RNA. J. Biol. Chem. 253, 1738–1742 (1978)
Lundberg, U. & Altman, S. Processing of the precursor to the catalytic RNA subunit of RNase P from Escherichia coli. RNA 1, 327–334 (1995)
Ow, M. C. & Kushner, S. R. Initiation of tRNA maturation by RNase E is essential for cell viability in Escherichia coli. Genes Dev. 16, 1102–1115 (2002)
Li, Z., Pandit, S. & Deutscher, M. P. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 18, 2878–2885 (1999)
Kim, K.-S. & Lee, Y. Regulation of 6S RNA biogenesis by switching utilization of both sigma factors and endoribonucleases. Nucleic Acids Res. 32, 6057–6068 (2004)
Bouvet, P. & Belasco, J. G. Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature 360, 488–491 (1992)
Mackie, G. A. Ribonuclease E is a 5′-end-dependent endonuclease. Nature 395, 720–723 (1998)
Mackie, G. A. Stabilization of circular rpsT mRNA demonstrates the 5′ end dependence of RNase E action in vivo. J. Biol. Chem. 275, 25069–25072 (2000)
Coburn, G. A. & Mackie, G. A. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. 62, 55–108 (1999)
Avarind, L. & Koonin, E. V. A natural classification of ribonucleases. Methods Enzymol. 341, 3–28 (2001)
McDowall, K. J. & Cohen, S. N. The N-terminal domain of the rne gene product has RNase E activity and is non-overlapping with the arginine-rich RNA binding site. J. Mol. Biol. 255, 349–355 (1996)
Kido, M. et al. RNase E polypeptides lacking a carboxyl-terminal half suppress a mukB mutation in Escherichia coli. J. Bacteriol. 178, 3917–3925 (1996)
McDowall, K. J., Hernandez, R. G., Lin-Chao, S. & Cohen, S. N. The ams-1 and rne-3071 temperature-sensitive mutations in the ams gene are in close proximity to each other and cause substitutions within a domain that resembles a product of the Escherichia coli mre locus. J. Bacteriol. 175, 4245–4249 (1993)
Wachi, M., Umitsuki, G., Shimizu, M., Takada, A. & Nagai, K. Escherichia coli cafA gene encodes a novel RNase, designated as RNase G, involved in processing of the 5′ end of 16S rRNA. Biochem. Biophys. Res. Commun. 259, 483–488 (1999)
Callaghan, A. J. et al. Studies of the RNA degradosome-organising domain of the Escherichia coli RNase E. J. Mol. Biol. 340, 965–979 (2004)
Carpousis, A. J. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem. Soc. Trans. 30, 150–155 (2002)
Callaghan, A. J. et al. The ‘Zn-link’: a metal-sharing interface that organizes the quaternary structure and catalytic site of the endoribonuclease, RNase E. Biochemistry 44, 4667–4675 (2005)
Lin-Chao, S. & Cohen, S. N. The rate of processing and degradation of antisense RNA I regulates the replication of ColE1-type plasmids in vivo. Cell 65, 1233–1242 (1991)
Redko, Y. et al. Determination of the catalytic parameters of the N-terminal half of E. coli ribonuclease E and the identification of critical functional groups in RNA substrates. J. Biol. Chem. 278, 44001–44008 (2003)
Jiang, X. & Belasco, J. G. Catalytic activation of multimeric RNase E and RNase G by 5′-monophosphorylated RNA. Proc. Natl Acad. Sci. USA 101, 9211–9216 (2004)
Kaberdin, V. R. Probing the substrate specificity of Escherichia coli RNase E using a novel oligonucleotide-based assay. Nucleic Acids Res. 31, 4710–4716 (2003)
Cohen, S. N. & McDowall, K. J. RNase E: still a wonderfully mysterious enzyme. Mol. Microbiol. 23, 1099–1106 (1997)
McDowall, K. J., Lin-Chao, S. & Cohen, S. N. A + U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J. Biol. Chem. 269, 10790–10796 (1994)
Lin-Chao, S., Wong, T. T., McDowall, K. J. & Cohen, S. N. Effects of nucleotide sequence on the specificity of rne-dependent and RNase E-mediated cleavages of RNA I encoded by the pBR322 plasmid. J. Biol. Chem. 269, 10797–10803 (1994)
Callaghan, A. J. et al. Quaternary structure and catalytic activity of the Escherichia coli ribonuclease E amino-terminal catalytic domain. Biochemistry 42, 13848–13855 (2003)
Acknowledgements
We thank the staff of the Daresbury Synchrotron Radiation Source, the European Synchrotron Radiation Facility and the Advanced Light Source synchrotrons for the use of facilities. We thank A. Murzin for pointing out the similarity with DNase I, and A. and T. Andreeva for helpful discussions about the structural deconvolution. We thank V. Chandran, K. Nagai, A. J. Carpousis and M. Symmons for advice. We thank C. Hill for synthesis of the protected RNA and L. Packman for protein and peptide analyses. This work was supported by the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The coordinates and structure factors have been deposited in the Protein Data Bank (PDB ID 2BX2, R2BX2SF and 2COB). Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Data
This file contains additional commentary on the structural features with references for all the supplementary subsections. (DOC 46 kb)
Supplementary Methods
Contains details of the crystallographic data collection, structure solution, refinement and analysis. (DOC 32 kb)
Supplementary Figure Legends
This contains the figure legends for Supplementary Figures S1–S3. (DOC 32 kb)
Supplementary Table 1
Contains the summary of the catalytic and RNA binding assays for RNase E catalytic domain mutants. (DOC 33 kb)
Supplementary Table 2
Crystallographic data (equivalent to the Nature formula Xray.doc) (DOC 85 kb)
Supplementary Table 3
A comprehensive list of mutagenic primers for RNase E catalytic domain mutants. (DOC 32 kb)
Supplementary Figure 1
Alignment of the RNase E catalytic domain homologues (PDF 298 kb)
Supplementary Figure 2
A figure showing the folds within the RNase E protomer. (PDF 810 kb)
Supplementary Figure 3
Showing the superposition of three RNase E/RNA complexes. (PDF 668 kb)
Rights and permissions
About this article
Cite this article
Callaghan, A., Marcaida, M., Stead, J. et al. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature 437, 1187–1191 (2005). https://doi.org/10.1038/nature04084
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature04084
This article is cited by
-
Heterologous Expression, Purification and Structural Characterization of Ribonuclease E from Mycobacterium smegmatis
Iranian Journal of Science (2023)
-
Relaxed Cleavage Specificity of Hyperactive Variants of Escherichia coli RNase E on RNA I
Journal of Microbiology (2023)
-
Tailoring the evolution of BL21(DE3) uncovers a key role for RNA stability in gene expression toxicity
Communications Biology (2021)
-
BiPSim: a flexible and generic stochastic simulator for polymerization processes
Scientific Reports (2021)
-
Trans-acting regulators of ribonuclease activity
Journal of Microbiology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.