Abstract
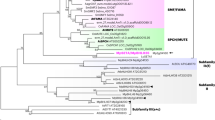
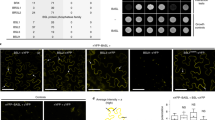
Stomata consist of a pair of guard cells that mediate gas and water-vapour exchange between plants and the atmosphere. Stomatal precursor cells—meristemoids—possess a transient stem-cell-like property and undergo several rounds of asymmetric divisions before further differentiation. Here we report that the Arabidopsis thaliana basic helix–loop–helix (bHLH) protein MUTE is a key switch for meristemoid fate transition. In the absence of MUTE, meristemoids abort after excessive asymmetric divisions and fail to differentiate stomata. Constitutive overexpression of MUTE directs the entire epidermis to adopt guard cell identity. MUTE has two paralogues: FAMA, a regulator of guard cell morphogenesis, and SPEECHLESS (SPCH). We show that SPCH directs the first asymmetric division that initiates stomatal lineage. Together, SPCH, MUTE and FAMA bHLH proteins control stomatal development at three consecutive steps: initiation, meristemoid differentiation and guard cell morphogenesis. Our findings highlight the roles of closely related bHLHs in cell type differentiation in plants and animals.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Nadeau, J. A. & Sack, F. D. Stomatal development in Arabidopsis. In The Arabidopsis Book (eds Somerville, C. & Meyerowitz, E. M.) doi:10.1199/tab.0066 (American Society of Plant Biologists, 2002)
Hetherington, A. M. & Woodward, F. I. The role of stomata in sensing and driving environmental change. Nature 424, 901–908 (2003)
Gray, J. E. et al. The HIC signalling pathway links CO2 perception to stomatal development. Nature 408, 713–716 (2000)
Berger, D. & Altmann, T. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14, 1119–1131 (2000)
von Groll, U., Berger, D. & Altmann, T. The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14, 1527–1539 (2002)
Nadeau, J. A. & Sack, F. D. Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697–1700 (2002)
Bergmann, D. C., Lukowitz, W. & Somerville, C. R. Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494–1497 (2004)
Shpak, E. D., McAbee, J. M., Pillitteri, L. J. & Torii, K. U. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309, 290–293 (2005)
Lai, L. B. et al. The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell 17, 2754–2767 (2005)
Poethig, R. S. EnhancerTraps 〈http://enhancertraps.bio.upenn.edu/〉 (2006)
Geisler, M. J., Deppong, D. O., Nadeau, J. A. & Sack, F. D. Stomatal neighbor cell polarity and division in Arabidopsis. Planta 216, 571–579 (2003)
Toledo-Ortiz, G., Huq, E. & Quail, P. H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749–1770 (2003)
Murre, C., McCaw, P. S. & Baltimore, D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56, 777–783 (1989)
Bailey, P. C. et al. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15, 2497–2502 (2003)
Buck, M. J. & Atchley, W. R. Phylogenetic analysis of plant basic helix-loop-helix proteins. J. Mol. Evol. 56, 742–750 (2003)
Heim, M. A. et al. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747 (2003)
Doe, C. Q. & Bowerman, B. Asymmetric cell division: fly neuroblast meets worm zygote. Curr. Opin. Cell Biol. 13, 68–75 (2001)
Ikeshima-Kataoka, H., Skeath, J. B., Nabeshima, Y., Doe, C. Q. & Matsuzaki, F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature 390, 625–629 (1997)
MacAlister, C. A., Ohashi-Ito, K & Bergmann, D. C. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature doi:10.1038/nature05491 (this issues)
Dengler, N. The developmental basis of anisophylly in Selaginella martensii. II. Histogenesis. Am. J. Bot. 70, 193–206 (1983)
Brown, R. & Lemmon, B. Development of stomata in Selaginella: Division polarity and plastid movements. Am. J. Bot. 72, 1914–1925 (1985)
Weintraub, H. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell 75, 1241–1244 (1993)
Olson, E. N. MyoD family: a paradigm for development? Genes Dev. 4, 1454–1461 (1990)
Campuzano, S. & Modolell, J. Patterning of the Drosophila nervous system: the achaete-scute gene complex. Trends Genet. 8, 202–208 (1992)
Olson, E. N. Interplay between proliferation and differentiation within the myogenic lineage. Dev. Biol. 154, 261–272 (1992)
Acknowledgements
We thank C. Doe, B. Wakimoto, J. McAbee, and T. Kakimoto for commenting on the manuscript; F. Sack and T. Altmann for a gift of flp, sdd1 and TMM::TMM–GFP; S. Poethig and D. Bergmann for E994; T. Nakagawa and ABRC for cloning vectors; SIGnAL and ABRC for T-DNA insertion lines; and P. Chan for confocal microscopy expertise. Our thanks to D. Bergmann for sharing unpublished results and discussion about stomatal development. This work was supported in part by the grants from DOE and NSF to K.U.T. L.J.P. was supported by the NRI USDA/CSREES fellowship, and K.U.T. is a CREST JST investigator.
Author Contributions K.U.T. supervised the entire project. K.U.T. and L.J.P. conceived and designed the experiments, and wrote the manuscript with comments from co-authors. L.J.P. isolated the mute mutant and performed characterization of mutants and transgenic plants with N.L.B. D.B.S. identified the MUTE gene by map-based cloning.
The NCBI/GenBank accession numbers for the genes described in this manuscript are: DQ863645 (MUTE mRNA), DQ864972 (MUTE genomic) and DQ868373 (SPCH mRNA).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Figures 1-6 and additional references. (PDF 4761 kb)
Supplementary Table 1
This file contains Supplementary Table 1 which shows PCR markers and their primer DNA sequence for genotypic analysis of stomatal development mutants. (XLS 18 kb)
Supplementary Table 2
This file contains Supplementary Table 2 which shows CAPS and dCAPS PCR markers and their primer DNA sequence used for map-based cloning of MUTE. (XLS 200 kb)
Supplementary Table 3a
This file contains Supplementary Table 3 which shows PCR primers and DNA sequence used for plasmid construction. (XLS 9 kb)
Supplementary Table 3b
This file contains Supplementary Table 3 which shows PCR primers and DNA sequence used for RT-PCR.. (XLS 16 kb)
Rights and permissions
About this article
Cite this article
Pillitteri, L., Sloan, D., Bogenschutz, N. et al. Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501–505 (2007). https://doi.org/10.1038/nature05467
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05467
This article is cited by
-
Time series single-cell transcriptional atlases reveal cell fate differentiation driven by light in Arabidopsis seedlings
Nature Plants (2023)
-
Stomatal regulators are co-opted for seta development in the astomatous liverwort Marchantia polymorpha
Nature Plants (2023)
-
Integration of light and ABA signaling pathways to combat drought stress in plants
Plant Cell Reports (2023)
-
Targeted expression of bgl23-D, a dominant-negative allele of ATCSLD5, affects cytokinesis of guard mother cells and exine formation of pollen in Arabidopsis thaliana
Planta (2023)
-
Genome-Wide Identification, In Silico Characterization of AtCOP1-Targeting Regulatory Proteins Network and their Expression Profiling in The COP1 Downregulated Arabidopsis thaliana
Journal of Plant Growth Regulation (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.