Abstract
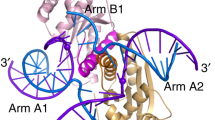
The four-way (Holliday) DNA junction is the central intermediate in homologous recombination, a ubiquitous process that is important in DNA repair and generation of genetic diversity1. The penultimate stage of recombination requires resolution of the DNA junction into nicked-duplex species by the action of a junction-resolving enzyme, examples of which have been identified in a wide variety of organisms2. These enzymes are nucleases that are highly selective for the structure of branched DNA. The mechanism of this selectivity has, however, been unclear in the absence of structural data. Here we present the crystal structure of the junction-resolving enzyme phage T7 endonuclease I in complex with a synthetic four-way DNA junction. Although the enzyme is structure-selective, significant induced fit occurs in the interaction, with changes in the structure of both the protein and the junction. The dimeric enzyme presents two binding channels that contact the backbones of the junction’s helical arms over seven nucleotides. These interactions effectively measure the relative orientations and positions of the arms of the junction, thereby ensuring that binding is selective for branched DNA that can achieve this geometry.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Holliday, R. A mechanism for gene conversion in fungi. Genet. Res. 5, 282–304 (1964)
Lilley, D. M. & White, M. F. The junction-resolving enzymes. Nature Rev. Mol. Cell Biol. 2, 433–443 (2001)
Center, M. S. & Richardson, C. C. An endonuclease induced after infection of Escherichia coli with bacteriophage T7. I. Purification and properties of the enzyme. J. Biol. Chem. 245, 6285–6291 (1970)
Sadowski, P. D. Bacteriophage T7 endonuclease. I. Properties of the enzyme purified from T7 phage-infected Escherichia coli B. J. Biol. Chem. 246, 209–216 (1971)
de Massey, B., Studier, F. W., Dorgai, L., Appelbaum, F. & Weisberg, R. A. Enzymes and the sites of genetic recombination: studies with gene-3 endonuclease of phage T7 and with site-affinity mutants of phage. Cold Spring Harb. Symp. Quant. Biol. 49, 715–726 (1984)
Lilley, D. M. & White, M. F. Resolving the relationships of resolving enzymes. Proc. Natl Acad. Sci. USA 97, 9351–9353 (2000)
Hadden, J. M., Convery, M. A., Déclais, A. C., Lilley, D. M. & Phillips, S. E. Crystal structure of the Holliday junction resolving enzyme T7 endonuclease I. Nature Struct. Biol. 8, 62–67 (2001)
Déclais, A.-C., Liu, J., Freeman, A. D. & Lilley, D. M. Structural recognition between a four-way DNA junction and a resolving enzyme. J. Mol. Biol. 359, 1261–1276 (2006)
Duckett, D. R., Giraud-Panis, M.-E. & Lilley, D. M. Binding of the junction-resolving enzyme bacteriophage T7 endonuclease I to DNA: separation of binding and catalysis by mutation. J. Mol. Biol. 246, 95–107 (1995)
Hadden, J. M., Déclais, A.-C., Phillips, S. E. & Lilley, D. M. Metal ions bound at the active site of the junction-resolving enzyme T7 endonuclease I. EMBO J. 21, 3505–3515 (2002)
Newman, M. et al. Crystal structure of restriction endonuclease BglI bound to its interrupted DNA recognition sequence. EMBO J. 17, 5466–5476 (1998)
Déclais, A.-C. et al. The complex between a four-way DNA junction and T7 endonuclease I. EMBO J. 22, 1398–1409 (2003)
Ortiz-Lombardia, M. et al. Crystal structure of a DNA Holliday junction. Nature Struct. Biol. 6, 913–917 (1999)
Eichman, B. F., Vargason, J. M., Mooers, B. H. M. & Ho, P. S. The Holliday junction in an inverted repeat DNA sequence: sequence effects on the structure of four-way junctions. Proc. Natl Acad. Sci. USA 97, 3971–3976 (2000)
Pöhler, J. R., Giraud-Panis, M. J. & Lilley, D. M. T4 endonuclease VII selects and alters the structure of the four-way DNA junction; binding of a resolution-defective mutant enzyme. J. Mol. Biol. 260, 678–696 (1996)
Biertümpfel, C. & Yang, W. &. Suck, D. Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature advance online publication, doi: 10.1038/nature06152 (2007)
Gopaul, D. N., Guo, F. & Van Duyne, G. D. Structure of the Holliday junction intermediate in Cre–loxP site-specific recombination. EMBO J. 17, 4175–4187 (1998)
Chen, Y., Narendra, U., Iype, L. E., Cox, M. M. & Rice, P. A. Crystal structure of a Flp recombinase–Holliday junction complex: assembly of an active oligomer by helix swapping. Mol. Cell 6, 885–897 (2000)
Ariyoshi, M., Nishino, T., Iwasaki, H., Shinagawa, H. & Morikawa, K. Crystal structure of the Holliday junction DNA in complex with a single RuvA tetramer. Proc. Natl Acad. Sci. USA 97, 8257–8262 (2000)
Roe, S. M. et al. Crystal structure of an octameric RuvA–Holliday junction complex. Mol. Cell. 2, 361–372 (1998)
Hargreaves, D. et al. Crystal structure of E. coli RuvA with bound DNA Holliday junction at 6 Å resolution. Nature Struct. Biol. 5, 441–446 (1998)
Bennett, R. J. & West, S. C. Structural analysis of the RuvC–Holliday junction complex reveals an unfolded junction. J. Mol. Biol. 252, 213–226 (1995)
White, M. F. & Lilley, D. M. The resolving enzyme CCE1 of yeast opens the structure of the four-way DNA junction. J. Mol. Biol. 266, 122–134 (1997)
Liu, J., Déclais, A.-C. & Lilley, D. M. Mechanistic aspects of the DNA junction-resolving enzyme T7 endonuclease I. Biochemistry 45, 3934–3942 (2006)
Bhattacharyya, A. et al. Model for the interaction of DNA junctions and resolving enzymes. J. Mol. Biol. 221, 1191–1207 (1991)
Parkinson, M. J. & Lilley, D. M. The junction-resolving enzyme T7 endonuclease I: quaternary structure and interaction with DNA. J. Mol. Biol. 270, 169–178 (1997)
McKinney, S. A., Déclais, A.-C., Lilley, D. M. & Ha, T. Structural dynamics of individual Holliday junctions. Nature Struct. Biol. 10, 93–97 (2003)
Joo, C., McKinney, S. A., Lilley, D. M. & Ha, T. Exploring rare conformational species and ionic effects in DNA Holliday junctions using single-molecule spectroscopy. J. Mol. Biol. 341, 739–751 (2004)
Collaborative computational project number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994)
Déclais, A. C., Liu, J., Freeman, A. D. & Lilley, D. M. Structural recognition between a four-way DNA junction and a resolving enzyme. J. Mol. Biol. 359, 1261–1276 (2006)
Leslie, A. G. W. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsl. Protein Crystallogr. 26, (1992)
Evans, P. R. SCALA. Joint CCP4 + ESF-EAMCB Newsl. Protein Crystallogr. 33, 22–24 (1997)
Storoni, L. C., McCoy, A. J. & Read, R. J. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D 60, 432–438 (2004)
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993)
Acknowledgements
We thank D. Flot at the ESRF for his assistance with the X-ray data collection and V. Sergeant for technical support. This work was supported by funds from the Wellcome Trust, BBSRC and Cancer Research UK.
Author Contributions J.M.H. purified and crystallized the complex, collected data, and built and refined the structure. A.-C.D. designed DNA oligonucleotides for co-crystallization and performed the molecular biology and biochemical studies. S.B.C. collected data and refined the structure of the complex. D.M.J.L. and S.E.V.P. conceived and designed the study and also helped to analyse the data. All authors discussed the results and wrote the paper.
Atomic coordinates and structure factors have been deposited in the PDB database under the accession code 2PFJ.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Tables 1-3, Supplementary Figure 1 with Legend and legends to Supplementary Videos 1 and 2. (PDF 596 kb)
Supplementary Video 1
The file contains Supplementary Video 1 showing rotating view of the Endo I binding site, formed by an alignment of the major grooves of DNA helices within the complex. (MOV 13821 kb)
Supplementary Video 2
The file contains Supplementary Video 2 depicting the re-orientation of the protein domains on binding the 4-way DNA junction. (MOV 3470 kb)
Rights and permissions
About this article
Cite this article
Hadden, J., Déclais, AC., Carr, S. et al. The structural basis of Holliday junction resolution by T7 endonuclease I. Nature 449, 621–624 (2007). https://doi.org/10.1038/nature06158
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06158
This article is cited by
-
An intercellular transfer of telomeres rescues T cells from senescence and promotes long-term immunological memory
Nature Cell Biology (2022)
-
Search and processing of Holliday junctions within long DNA by junction-resolving enzymes
Nature Communications (2022)
-
Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1
Nature Communications (2020)
-
Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase
Scientific Reports (2020)
-
Structural basis of sequence-specific Holliday junction cleavage by MOC1
Nature Chemical Biology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.