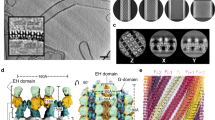
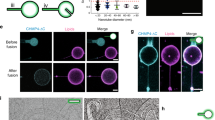
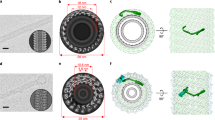
Abstract
The ability to actively remodel membranes in response to nucleotide hydrolysis has largely been attributed to GTPases of the dynamin superfamily, and these have been extensively studied1. Eps15 homology (EH)-domain-containing proteins (EHDs/RME-1/pincher) comprise a less-well-characterized class of highly conserved eukaryotic ATPases implicated in clathrin-independent endocytosis2, and recycling from endosomes3,4. Here we show that EHDs share many common features with the dynamin superfamily, such as a low affinity for nucleotides, the ability to tubulate liposomes in vitro, oligomerization around lipid tubules in ring-like structures and stimulated nucleotide hydrolysis in response to lipid binding. We present the structure of EHD2, bound to a non-hydrolysable ATP analogue, and provide evidence consistent with a role for EHDs in nucleotide-dependent membrane remodelling in vivo. The nucleotide-binding domain is involved in dimerization, which creates a highly curved membrane-binding region in the dimer. Oligomerization of dimers occurs on another interface of the nucleotide-binding domain, and this allows us to model the EHD oligomer. We discuss the functional implications of the EHD2 structure for understanding membrane deformation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
18 October 2007
The AOP version of this paper contained the term 'Epsin homology (EH)-domain...'. This was corrected to 'Eps15 homology (EH)-domain...' in the 18 October issue
References
Praefcke, G. J. & McMahon, H. T. The dynamin superfamily: universal membrane tubulation and fission molecules? Nature Rev. Mol. Cell Biol. 5, 133–147 (2004)
Shao, Y. et al. Pincher, a pinocytic chaperone for nerve growth factor/TrkA signaling endosomes. J. Cell Biol. 157, 679–691 (2002)
Grant, B. et al. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nature Cell Biol. 3, 573–579 (2001)
Caplan, S. et al. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 21, 2557–2567 (2002)
Blume, J. J., Halbach, A., Behrendt, D., Paulsson, M. & Plomann, M. EHD proteins are associated with tubular and vesicular compartments and interact with specific phospholipids. Exp. Cell Res. 313, 219–231 (2007)
George, M. et al. Shared as well as distinct roles of EHD proteins revealed by biochemical and functional comparisons in mammalian cells and C. elegans . BMC Cell Biol. 8, 3 (2007)
Jovic, M., Naslavsky, N., Rapaport, D., Horowitz, M. & Caplan, S. EHD1 regulates β1 integrin endosomal transport: effects on focal adhesions, cell spreading and migration. J. Cell Sci. 120, 802–814 (2007)
Lin, S. X., Grant, B., Hirsh, D. & Maxfield, F. R. Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nature Cell Biol. 3, 567–572 (2001)
Park, S. Y. et al. EHD2 interacts with the insulin-responsive glucose transporter (GLUT4) in rat adipocytes and may participate in insulin-induced GLUT4 recruitment. Biochemistry 43, 7552–7562 (2004)
Rotem-Yehudar, R., Galperin, E. & Horowitz, M. Association of insulin-like growth factor 1 receptor with EHD1 and SNAP29. J. Biol. Chem. 276, 33054–33060 (2001)
Valdez, G. et al. Pincher-mediated macroendocytosis underlies retrograde signaling by neurotrophin receptors. J. Neurosci. 25, 5236–5247 (2005)
Braun, A. et al. EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Mol. Biol. Cell 16, 3642–3658 (2005)
Lee, D. W. et al. ATP binding regulates oligomerization and endosome association of RME-1 family proteins. J. Biol. Chem. 280, 17213–17220 (2005)
Guilherme, A. et al. EHD2 and the novel EH domain binding protein EHBP1 couple endocytosis to the actin cytoskeleton. J. Biol. Chem. 279, 10593–10605 (2004)
Ghosh, A., Praefcke, G. J., Renault, L., Wittinghofer, A. & Herrmann, C. How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature 440, 101–104 (2006)
Reubold, T. F. et al. Crystal structure of the GTPase domain of rat dynamin 1. Proc. Natl Acad. Sci. USA 102, 13093–13098 (2005)
Low, H. H. & Lowe, J. A bacterial dynamin-like protein. Nature 444, 766–769 (2006)
de Beer, T., Carter, R. E., Lobel-Rice, K. E., Sorkin, A. & Overduin, M. Structure and Asn-Pro-Phe binding pocket of the Eps15 homology domain. Science 281, 1357–1360 (1998)
de Beer, T. et al. Molecular mechanism of NPF recognition by EH domains. Nature Struct. Biol. 7, 1018–1022 (2000)
Marks, B. et al. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature 410, 231–235 (2001)
Martens, S., Kozlov, M. M. & McMahon, H. T. How synaptotagmin promotes membrane fusion. Science 316, 1205–1208 (2007)
Zimmerberg, J. & Kozlov, M. M. How proteins produce cellular membrane curvature. Nature Rev. Mol. Cell Biol. 7, 9–19 (2006)
Lenzen, C., Cool, R. H. & Wittinghofer, A. Analysis of intrinsic and CDC25-stimulated guanine nucleotide exchange of p21ras-nucleotide complexes by fluorescence measurements. Methods Enzymol. 255, 95–109 (1995)
Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993)
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell vonstants. J. of Appl. Crystallogr. 26, 795–800 (1993)
Sheldrick, G. M. & Schneider, T. R. SHELXL: High-resolution refinement. Methods Enzymol. 277, 319–343 (1997)
de la Fortelle, E. & Bricogne, G. in Methods in Enzymology (eds Carter, C. W. Jr & Sweet, R. M.). 472–494 (1997)
McRee, D. E. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125, 156–65 (1999)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240 (1997)
Laskowski, R. A., Macarthur, M. W., Moss, D. S. & Thornton, J. M. Procheck—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993)
Kraulis, P. J. Molscript—a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991)
Merritt, E. A. & Murphy, M. E. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D 50, 869–73 (1994)
Landau, M. et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, W299–W302 (2005)
Potterton, E., McNicholas, S., Krissinel, E., Cowtan, K. & Noble, M. The CCP4 molecular graphics project. Acta Crystallogr. D 58, 1955–1957 (2002)
DeLano, W. L. The PyMOL Molecular Graphics System (DeLano Scientific, Palo Alto, California, USA, 2002)
Guex, N. & Peitsch, M. C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophor. 18, 2714–2723 (1997)
Collaborative Computational Project The CCP4 suite: programs for protein crystallography. Acta Crystallogr . D 50, 760 (1994)
Philo, J. S. A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Analyt. Biochem. 279, 151–163 (2000)
Philo, J. S. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Analyt. Biochem. 354, 238–246 (2006)
Laue, T. M., Shah, B. D., Ridgeway, T. M. & Pelletier, S. L. in Analytical Ultracentrifugation in Biochemistry and Polymer Science (eds Harding, S. E., Rowe, A. J. & Horton, J. C.) 90–125 (Roy. Soc. of Chem., Cambridge, UK, 1992)
Acknowledgements
Long-term fellowships supported O.D. (The International Human Frontier Science Program Organization), R.L. (Swedish Research Council) and S.M. (EMBO). We thank M. Plomann for providing the complementary DNAs for mammalian EHDs, and the ESRF beam staff in Grenoble for their support. The authors declare no competing financial interests.
The atomic coordinates of mouse EHD2 have been deposited in the Protein Data Bank (PDB) with the accession number 2QPT.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information 1
This file contains Supplementary Figures 1-12 with Legends, Supplementary Table 1 with the data collection statistics and additional references. (PDF 2225 kb)
Supplementary Information 2
The file contains Supplementary Video 1 which shows EHD2 wild-type was over-expressed in HeLa cells for 24 h and imaged by EPI-fluorescence for approximately 30 min. Some of the tubules and puncta are dynamic. (MOV 4707 kb)
Supplementary Information 3
The file contains Supplementary Video 2 which shows cropped area of EHD2 WT (Video 1) at higher resolution. (MOV 5319 kb)
Supplementary Information 4
The file contains Supplementary Video 3 which shows EHD2 T94A was over-expressed in HeLa cells for 24 h and imaged by EPI-fluorescence for approximately 30 min. There are only tubules, and these are mostly static. Cropped movies show a smaller area at higher resolution. (MOV 6652 kb)
Supplementary Information 5
The file contains Supplementary Video 4 which shows cropped area of EHD2 T94A (Video 2) at higher resolution. (MOV 7726 kb)
Supplementary Information 6
The file contains Supplementary Video 5 which shows EHD2 I157Q was over-expressed in HeLa cells for 24h and imaged by EPI-fluorescence for approximately 30min. No tubules can be found and the puncta are mostly motile. (MOV 9013 kb)
Supplementary Information 7
The file contains Supplementary Video 6 which shows cropped area of EHD2 I157Q (Video 4) at higher resolution. (MOV 6353 kb)
Supplementary Information 8
The file contains Supplementary Data with PDB coordinates of the proposed EHD2 oligomer. Four EHD2 dimers (in the absence of the EH domain) were aligned as described in Methods. All lipid interaction sites point towards the putative membrane interface. Molecules B and C which have been used for the initial alignment with GBP1 are related via a 2-fold axis and the nucleotides of these molecules are oriented in a head-to-head fashion. (ZIP 455 kb)
Rights and permissions
About this article
Cite this article
Daumke, O., Lundmark, R., Vallis, Y. et al. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature 449, 923–927 (2007). https://doi.org/10.1038/nature06173
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06173
This article is cited by
-
EHD1-dependent traffic of IGF-1 receptor to the cell surface is essential for Ewing sarcoma tumorigenesis and metastasis
Communications Biology (2023)
-
Structural mechanism of mitochondrial membrane remodelling by human OPA1
Nature (2023)
-
The molecular organization of differentially curved caveolae indicates bendable structural units at the plasma membrane
Nature Communications (2022)
-
Insights into Membrane Curvature Sensing and Membrane Remodeling by Intrinsically Disordered Proteins and Protein Regions
The Journal of Membrane Biology (2022)
-
Lysine acetylation regulates the interaction between proteins and membranes
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.