Abstract
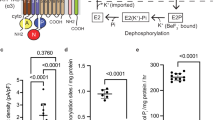
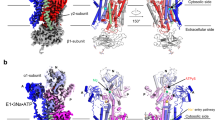
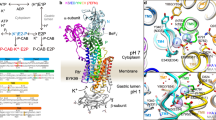
The Na+,K+-ATPase generates electrochemical gradients for sodium and potassium that are vital to animal cells, exchanging three sodium ions for two potassium ions across the plasma membrane during each cycle of ATP hydrolysis. Here we present the X-ray crystal structure at 3.5 Å resolution of the pig renal Na+,K+-ATPase with two rubidium ions bound (as potassium congeners) in an occluded state in the transmembrane part of the α-subunit. Several of the residues forming the cavity for rubidium/potassium occlusion in the Na+,K+-ATPase are homologous to those binding calcium in the Ca2+-ATPase of sarco(endo)plasmic reticulum. The β- and γ-subunits specific to the Na+,K+-ATPase are associated with transmembrane helices αM7/αM10 and αM9, respectively. The γ-subunit corresponds to a fragment of the V-type ATPase c subunit. The carboxy terminus of the α-subunit is contained within a pocket between transmembrane helices and seems to be a novel regulatory element controlling sodium affinity, possibly influenced by the membrane potential.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Skou, J. C. The influence of some cations on an adenosine triphosphatase from peripheral nerves. Biochim. Biophys. Acta 1000, 439–446 (1957)
Post, R. L., Hegyvary, C. & Kume, S. Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J. Biol. Chem. 247, 6530–6540 (1972)
Glynn, I. M. Annual review prize lecture. 'All hands to the sodium pump'. J. Physiol. (Lond.) 462, 1–30 (1993)
Gadsby, D. C., Rakowski, R. F. & De Weer, P. Extracellular access to the Na,K Pump: pathway similar to ion channel. Science 260, 100–103 (1993)
Apell, H. J. & Karlish, S. J. Functional properties of Na,K-ATPase, and their structural implications, as detected with biophysical techniques. J. Membr. Biol. 180, 1–9 (2001)
Geering, K. The functional role of β subunits in oligomeric P-type ATPases. J. Bioenerg. Biomembr. 33, 425–438 (2001)
Lutsenko, S. & Kaplan, J. H. An essential role for the extracellular domain of the Na,K-ATPase β-subunit in cation occlusion. Biochemistry 32, 6737–6743 (1993)
Garty, H. & Karlish, S. J. Role of FXYD proteins in ion transport. Annu. Rev. Physiol. 68, 431–459 (2006)
Therien, A. G. & Blostein, R. Mechanisms of sodium pump regulation. Am. J. Physiol. Cell Physiol. 279, C541–C566 (2000)
Toyoshima, C., Nakasako, M., Nomura, H. & Ogawa, H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature 405, 647–655 (2000)
Sorensen, T. L., Moller, J. V. & Nissen, P. Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science 304, 1672–1675 (2004)
Toyoshima, C., Nomura, H. & Tsuda, T. Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature 432, 361–368 (2004)
Olesen, C., Sorensen, T. L., Nielsen, R. C., Moller, J. V. & Nissen, P. Dephosphorylation of the calcium pump coupled to counterion occlusion. Science 306, 2251–2255 (2004)
Vilsen, B., Andersen, J. P., Petersen, J. & Jørgensen, P. L. Occlusion of 22Na+ and 86Rb+ in membrane-bound and soluble protomeric αβ-units of Na,K-ATPase. J. Biol. Chem. 262, 10511–10517 (1987)
Danko, S., Yamasaki, K., Daiho, T. & Suzuki, H. Distinct natures of beryllium fluoride-bound, aluminum fluoride-bound, and magnesium fluoride-bound stable analogues of an ADP-insensitive phosphoenzyme intermediate of sarcoplasmic reticulum Ca2+-ATPase: changes in catalytic and transport sites during phosphoenzyme hydrolysis. J. Biol. Chem. 279, 14991–14998 (2004)
Vilsen, B. & Andersen, J. P. Mutation to the glutamate in the fourth membrane segment of Na+,K+- ATPase and Ca2+-ATPase affects cation binding from both sides of the membrane and destabilizes the occluded enzyme forms. Biochemistry 37, 10961–10971 (1998)
Einholm, A. P., Andersen, J. P. & Vilsen, B. Importance of Leu99 in transmembrane segment M1 of the Na+,K+-ATPase in the binding and occlusion of K+ . J. Biol. Chem. 282, 23854–23866 (2007)
Vilsen, B. Mutant Glu781Ala of the rat kidney Na+,K+-ATPase displays low cation affinity and catalyzes ATP hydrolysis at a high rate in the absence of potassium ions. Biochemistry 34, 1455–1463 (1995)
Kuntzweiler, T. A., Arguello, J. M. & Lingrel, J. B. Asp804 and Asp808 in the transmembrane domain of the Na,K-ATPase α subunit are cation coordinating residues. J. Biol. Chem. 271, 29682–29687 (1996)
Blostein, R., Wilczynska, A., Karlish, S. J., Arguello, J. M. & Lingrel, J. B. Evidence that Ser775 in the α subunit of the Na,K-ATPase is a residue in the cation binding pocket. J. Biol. Chem. 272, 24987–24993 (1997)
Pedersen, P. A., Nielsen, J. M., Rasmussen, J. H. & Jorgensen, P. L. Contribution to Tl+, K+, and Na+ binding of Asn776, Ser775, Thr774, Thr772, and Tyr771 in cytoplasmic part of fifth transmembrane segment in α-subunit of renal Na,K-ATPase. Biochemistry 37, 17818–17827 (1998)
Olesen, C. et al. The structural basis of calcium transport by the calcium pump. Nature doi: 10.1038/nature06418. (this issue)
Clarke, D. M., Loo, T. W., Inesi, G. & MacLennan, D. H. Location of high affinity Ca2+-binding sites within the predicted transmembrane domain of the sarcoplasmic reticulum Ca2+-ATPase. Nature 339, 476–478 (1989)
Andersen, J. P. & Vilsen, B. Amino acids Asn796 and Thr799 of the Ca2+-ATPase of sarcoplasmic reticulum bind Ca2+ at different sites. J. Biol. Chem. 269, 15931–15936 (1994)
Jardetzky, O. Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966)
Donnet, C., Arystarkhova, E. & Sweadner, K. J. Thermal denaturation of the Na,K-ATPase provides evidence for α-α oligomeric interaction and γ subunit association with the C-terminal domain. J. Biol. Chem. 276, 7357–7365 (2001)
Colonna, T. E., Huynh, L. & Fambrough, D. M. Subunit interactions in the Na,K-ATPase explored with the yeast two-hybrid system. J. Biol. Chem. 272, 12366–12372 (1997)
Rice, W. J., Young, H. S., Martin, D. W., Sachs, J. R. & Stokes, D. L. Structure of the Na+,K+-ATPase at 11-Å resolution: comparison with Ca2+-ATPase in E1 and E2 states. Biophys. J. 80, 2187–2197 (2001)
Hebert, H., Purhonen, P., Vorum, H., Thomsen, K. & Maunsbach, A. B. Three-dimensional structure of renal Na,K-ATPase from cryo-electron microscopy of two-dimensional crystals. J. Mol. Biol. 314, 478–494 (2001)
Murata, T., Yamato, I., Kakinuma, Y., Leslie, A. G. & Walker, J. E. Structure of the rotor of the V-Type Na+-ATPase from Enterococcus hirae . Science 308, 654–659 (2005)
Teriete, P., Franzin, C. M., Choi, J. & Marassi, F. M. Structure of the Na,K-ATPase regulatory protein FXYD1 in micelles. Biochemistry 46, 6774–6783 (2007)
Li, C. et al. Structural and functional interactions sites between Na+,K+-ATPase and FXYD proteins. J. Biol. Chem. 279, 38895–38902 (2004)
Meij, I. C. et al. Dominant isolated renal magnesium loss is caused by misrouting of the Na+,K+-ATPase γ-subunit. Nature Genet. 26, 265–266 (2000)
Einholm, A. P., Toustrup-Jensen, M., Andersen, J. P. & Vilsen, B. Mutation of Gly-94 in transmembrane segment M1 of Na+,K+-ATPase interferes with Na+ and K+ binding in E2P conformation. Proc. Natl Acad. Sci. USA 102, 11254–11259 (2005)
Pedersen, B. P., Buch-Pedersen, M. J., Morth, J. P., Palmgren, M. G. & Nissen, P. Crystal structure of a plasma membrane proton pump. doi: 10/1038/nature06417 Nature. (this issue)
Håkansson, K. O. The crystallographic structure of Na,K-ATPase N-domain at 2.6 Å resolution. J. Mol. Biol. 332, 1175–1182 (2003)
Hilge, M. et al. ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase. Nature Struct. Biol. 10, 468–474 (2003)
Sweadner, K. J. & Feschenko, M. S. Predicted location and limited accessibility of protein kinase A phosphorylation site on Na-K-ATPase. Am. J. Physiol. Cell Physiol. 280, C1017–C1026 (2001)
Vilsen, B., Ramlov, D. & Andersen, J. P. Functional consequences of mutations in the transmembrane core region for cation translocation and energy transduction in the Na+,K+-ATPase and the SR Ca2+-ATPase. Ann. NY Acad. Sci. 834, 297–309 (1997)
Ogawa, H. & Toyoshima, C. Homology modeling of the cation binding sites of Na+K+-ATPase. Proc. Natl Acad. Sci. USA 99, 15977–15982 (2002)
Li, C., Capendeguy, O., Geering, K. & Horisberger, J. D. A third Na+-binding site in the sodium pump. Proc. Natl Acad. Sci. USA 102, 12706–12711 (2005)
Imagawa, T., Yamamoto, T., Kaya, S., Sakaguchi, K. & Taniguchi, K. Thr-774 (transmembrane segment M5), Val-920 (M8), and Glu-954 (M9) are involved in Na+ transport, and Gln-923 (M8) is essential for Na,K-ATPase activity. J. Biol. Chem. 280, 18736–18744 (2005)
Jiang, Y., Ruta, V., Chen, J., Lee, A. & MacKinnon, R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature 423, 42–48 (2003)
Bass, R. B., Strop, P., Barclay, M. & Rees, D. C. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science 298, 1582–1587 (2002)
Crambert, G. et al. Transport and pharmacological properties of nine different human Na, K-ATPase isozymes. J. Biol. Chem. 275, 1976–1986 (2000)
Jørgensen, P. L. Purification and characterization of (Na+ + K+)-ATPase. III. Purification from the outer medulla of mammalian kidney after selective removal of membrane components by SDS. Biochim. Biophys. Acta 356, 36–52 (1974)
Rodacker, V., Toustrup-Jensen, M. & Vilsen, B. Mutations Phe785Leu and Thr618Met in Na+,K+-ATPase, associated with familial rapid-onset dystonia parkinsonism, interfere with Na+ interaction by distinct mechanisms. J. Biol. Chem. 281, 18539–18548 (2006)
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993)
McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D 61, 458–464 (2005)
Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 (2006)
Cowtan, K. D. & Main, P. Phase combination and cross validation in iterated density-modification calculations. Acta Crystallogr. D 52, 43–48 (1996)
Terwilliger, T. C. Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 (2000)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Acknowledgements
We thank E. Pohl, C. Schulze-Briese, and T. Tomizaki for extensive assistance with synchrotron data collection and Anna Marie Nielsen, Jytte Jørgensen, Kirsten Lykke Pedersen, and Lene Jacobsen for technical assistance. We are thankful to Hanne Poulsen, Anja P. Einholm, and Laure Yatime for discussion, and to Dr. Otto Hansen, University of Aarhus, for the γ-subunit specific polyclonal antibody. This work was supported financially through the DANSYNC programme, the Centre for Structural Biology of the Danish Research Council for Natural Sciences, and a Hallas-Møller stipend from the Novo-Nordisk Foundation (to P.N.), as well as by research grants from the Lundbeck Foundation, the Novo Nordisk Foundation (Fabrikant Vilhelm Pedersen og Hustrus Legat), and the Danish Medical Research Council (to B.V.). B.P.P. is supported by a fellowship from the Aarhus Graduate School of Science.
Author Contributions J.P.M. performed crystallization experiments, data collection and processing, structure determination and refinement, and structural analysis. B.P.P. assisted in data collection, structure determination and analysis. T.L.S. identified initial crystallization conditions. M.S.T.J. produced mutant enzyme and characterized it functionally. J.P. performed occlusion experiments and prepared and characterized Na+,K+-ATPase samples as required for crystallization. P.N., B.V., and J.P.A. supervised the project and analysed the structure. The paper was written by J.P.M., P.N., B.V., and J.P.A., P.N. and B.V. have contributed equally and with equal resources to the project.
Author information
Authors and Affiliations
Corresponding authors
Additional information
The Na+,K+-ATPase has the PDB code 3B8E.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures S1-S6 with Legends. (PDF 5267 kb)
Supplementary Movie
This file contains Supplementary Movie 1 entitled ‘Rotating model of pig renal Na+,K+-ATPase’ The movie shows the α1 β1 γ complex of the Na+,K+-ATPase from pig kidney. The α-subunit is in blue, β in wheat, γ in red, and the C-terminal switch of α in light red. The two Rb+ ions in the membrane are indicated by magenta spheres, and the MgF42- and Mg2+ bound at the cytoplasmic P domain are indicated by orange and grey spheres, respectively. (SWF 9309 kb)
Rights and permissions
About this article
Cite this article
Morth, J., Pedersen, B., Toustrup-Jensen, M. et al. Crystal structure of the sodium–potassium pump. Nature 450, 1043–1049 (2007). https://doi.org/10.1038/nature06419
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature06419
This article is cited by
-
A century of exercise physiology: effects of muscle contraction and exercise on skeletal muscle Na+,K+-ATPase, Na+ and K+ ions, and on plasma K+ concentration—historical developments
European Journal of Applied Physiology (2024)
-
The effect of cytochrome c on Na,K-ATPase
Journal of Bioenergetics and Biomembranes (2024)
-
Identified and potential internalization signals involved in trafficking and regulation of Na+/K+ ATPase activity
Molecular and Cellular Biochemistry (2023)
-
Electrostatic switch mechanisms of membrane protein trafficking and regulation
Biophysical Reviews (2023)
-
Structure and function of H+/K+ pump mutants reveal Na+/K+ pump mechanisms
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.