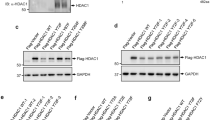
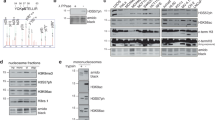
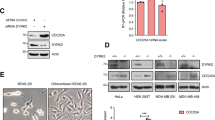
Abstract
Life and death fate decisions allow cells to avoid massive apoptotic death in response to genotoxic stress. Although the regulatory mechanisms and signalling pathways controlling DNA repair and apoptosis are well characterized, the precise molecular strategies that determine the ultimate choice of DNA repair and survival or apoptotic cell death remain incompletely understood. Here we report that a protein tyrosine phosphatase, EYA, is involved in promoting efficient DNA repair rather than apoptosis in response to genotoxic stress in mammalian embryonic kidney cells by executing a damage-signal-dependent dephosphorylation of an H2AX carboxy-terminal tyrosine phosphate (Y142). This post-translational modification determines the relative recruitment of either DNA repair or pro-apoptotic factors to the tail of serine phosphorylated histone H2AX (γ-H2AX) and allows it to function as an active determinant of repair/survival versus apoptotic responses to DNA damage, revealing an additional phosphorylation-dependent mechanism that modulates survival/apoptotic decisions during mammalian organogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kumar, J. P. Signalling pathways in Drosophila and vertebrate retinal development. Nature Rev. Genet. 2, 846–857 (2001)
Silver, S. J. & Rebay, I. Signaling circuitries in development: insights from the retinal determination gene network. Development 132, 3–13 (2005)
Pignoni, F. et al. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91, 881–891 (1997)
Bonini, N. M., Leiserson, W. M. & Benzer, S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell 72, 379–395 (1993)
Xu, P. X. et al. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nature Genet. 23, 113–117 (1999)
Li, X. et al. Eya protein phosphatase activity regulates Six1–Dach–Eya transcriptional effects in mammalian organogenesis. Nature 426, 247–254 (2003)
Rayapureddi, J. P. et al. Eyes absent represents a class of protein tyrosine phosphatases. Nature 426, 295–298 (2003)
Tootle, T. L. et al. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature 426, 299–302 (2003)
Rayapureddi, J. P. et al. Characterization of a plant, tyrosine-specific phosphatase of the aspartyl class. Biochemistry 44, 751–758 (2005)
Lu, C. et al. Cell apoptosis: requirement of H2AX in DNA ladder formation, but not for the activation of caspase-3. Mol. Cell 23, 121–132 (2006)
Bassing, C. H. & Alt, F. W. The cellular response to general and programmed DNA double strand breaks. DNA Repair 3, 781–796 (2004)
Bassing, C. H. et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl Acad. Sci. USA 99, 8173–8178 (2002)
Karagiannis, T. C. & El-Osta, A. Chromatin modifications and DNA double-strand breaks: the current state of play. Leukemia 21, 195–200 (2007)
van Attikum, H. & Gasser, S. M. The histone code at DNA breaks: a guide to repair? Nature Rev. Mol. Cell Biol. 6, 757–765 (2005)
Lee, Y. M. et al. Determination of hypoxic region by hypoxia marker in developing mouse embryos in vivo: a possible signal for vessel development. Dev. Dyn. 220, 175–186 (2001)
Haase, V. H. Hypoxia-inducible factors in the kidney. Am. J. Physiol. Renal Physiol. 291, F271–F281 (2006)
Fernandez-Capetillo, O. et al. H2AX: the histone guardian of the genome. DNA Repair 3, 959–967 (2004)
Rogakou, E. P. et al. Megabase chromatin domains involved in DNA double-strand breaks in vivo . J. Cell Biol. 146, 905–916 (1999)
Berkovich, E., Monnat, R. J. & Kastan, M. B. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nature Cell Biol. 9, 683–690 (2007)
Berkovich, E., Monnat, R. J. & Kastan, M. B. Assessment of protein dynamics and DNA repair following generation of DNA double-strand breaks at defined genomic sites. Nature Protocols 3, 915–922 (2008)
Matsuoka, S. et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 (2007)
Stokes, M. P. et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl Acad. Sci. USA 104, 19855–19860 (2007)
Lavin, M. F. & Kozlov, S. ATM activation and DNA damage response. Cell Cycle 6, 931–942 (2007)
Stucki, M. et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123, 1213–1226 (2005)
Lee, M. S. et al. Structure of the BRCT repeat domain of MDC1 and its specificity for the free COOH-terminal end of the γ-H2AX histone tail. J. Biol. Chem. 280, 32053–32056 (2005)
Kim, J. E., Minter-Dykhouse, K. & Chen, J. Signaling networks controlled by the MRN complex and MDC1 during early DNA damage responses. Mol. Carcinog. 45, 403–408 (2006)
Wu, X. et al. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405, 477–482 (2000)
Celeste, A. et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature Cell Biol. 5, 675–679 (2003)
Duilio, A. et al. A rat brain mRNA encoding a transcriptional activator homologous to the DNA binding domain of retroviral integrases. Nucleic Acids Res. 19, 5269–5274 (1991)
Minopoli, G. et al. Essential roles for Fe65, Alzheimer amyloid precursor-binding protein, in the cellular response to DNA damage. J. Biol. Chem. 282, 831–835 (2007)
Nakaya, T., Kawai, T. & Suzuki, T. Regulation of FE65 nuclear translocation and function by amyloid β-protein precursor in osmotically stressed cells. J. Biol. Chem. 283, 19119–19131 (2008)
Celeste, A. et al. Genomic instability in mice lacking histone H2AX. Science 296, 922–927 (2002)
Jemc, J. & Rebay, I. Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent. Dev. Biol. 310, 416–429 (2007)
Xiao, A. et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature 457, 57–62 (2009)
Sambrook, J. & Russell, D. W. Molecular Cloning: a Laboratory Manual 3rd edn (Cold Spring Harbor Laboratory Press, 2001)
Acknowledgements
We thank M. Kastan for providing reagents/technical assistance for the I-PpoI system. We thank V. Lunyak, J. Dixon and R. Koladner for review and discussions. We thank the laboratory of R. S. Johnson for use of equipment and advice on hypoxia incubations, as well as H. Taylor for animal care assistance and C. Nelson for cell culture assistance. We thank A. Nussenzweig, Y. Xu and H. Song for H2ax-/- MEFs. We thank J. Hightower and M. Fisher for assistance with figure and manuscript preparation. We additionally thank X. Li and W. Liu. M.G.R. is an HHMI Investigator. This work was supported by grants from NIH and NCI to M.G.R. and C.K.G. This work also was supported by the Sogang University Research Grant of 2008 to B.G.J and PCF and USAMRAA grants to M.G.R.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Figures 1-12 with Legends and Supplementary Table 1. (PDF 497 kb)
Rights and permissions
About this article
Cite this article
Cook, P., Ju, B., Telese, F. et al. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature 458, 591–596 (2009). https://doi.org/10.1038/nature07849
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07849
This article is cited by
-
Retinal determination gene networks: from biological functions to therapeutic strategies
Biomarker Research (2023)
-
Changes in calpain-2 expression during glioblastoma progression predisposes tumor cells to temozolomide resistance by minimizing DNA damage and p53-dependent apoptosis
Cancer Cell International (2023)
-
EYA4 promotes breast cancer progression and metastasis through its role in replication stress avoidance
Molecular Cancer (2023)
-
Ex vivo drug response heterogeneity reveals personalized therapeutic strategies for patients with multiple myeloma
Nature Cancer (2023)
-
Direct targeting of amplified gene loci for proapoptotic anticancer therapy
Nature Biotechnology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.