Abstract
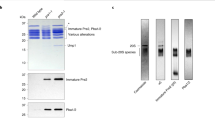
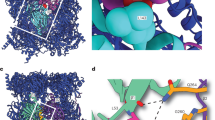
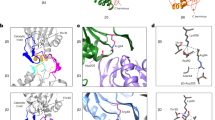
The proteasome is a protease that controls diverse processes in eukaryotic cells. Its regulatory particle (RP) initiates the degradation of ubiquitin–protein conjugates by unfolding the substrate and translocating it into the proteasome core particle (CP) to be degraded1. The RP has 19 subunits, and their pathway of assembly is not understood. Here we show that in the yeast Saccharomyces cerevisiae three proteins are found associated with RP but not with the RP–CP holoenzyme: Nas6, Rpn14 and Hsm3. Mutations in the corresponding genes confer proteasome loss-of-function phenotypes, despite their virtual absence from the holoenzyme. These effects result from deficient RP assembly. Thus, Nas6, Rpn14 and Hsm3 are RP chaperones. The RP contains six ATPases–the Rpt proteins–and each RP chaperone binds to the carboxy-terminal domain of a specific Rpt. We show in an accompanying study2 that RP assembly is templated through the Rpt C termini, apparently by their insertion into binding pockets in the CP. Thus, RP chaperones may regulate proteasome assembly by directly restricting the accessibility of Rpt C termini to the CP. In addition, competition between the RP chaperones and the CP for Rpt engagement may explain the release of RP chaperones as proteasomes mature.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 (2009)
Park, S. et al. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature 10.1038/nature08065 (this issue)
Leggett, D. S. et al. Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 (2002)
Verma, R. et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11, 3425–3439 (2000)
Wang, X. et al. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry 46, 3553–3565 (2007)
Dawson, S., Higashitsuji, H., Wilkinson, A. J., Fujita, J. & Mayer, R. J. Gankyrin: a new oncoprotein and regulator of pRb and p53. Trends Cell Biol. 16, 229–233 (2006)
Guerrero, C., Milenkovic, T., Przulj, N., Kaiser, P. & Huang, L. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc. Natl Acad. Sci. USA 105, 13333–13338 (2008)
Park, Y. et al. Proteasomal ATPase-associated factor 1 negatively regulates proteasome activity by interacting with proteasomal ATPases. Mol. Cell. Biol. 25, 3842–3853 (2005)
Lassot, I. et al. The proteasome regulates HIV-1 transcription by both proteolytic and nonproteolytic mechanisms. Mol. Cell 25, 369–383 (2007)
Collins, G. A. & Tansey, W. P. The proteasome: a utility tool for transcription? Curr. Opin. Genet. Dev. 16, 197–202 (2006)
Imai, J., Maruya, M., Yashiroda, H., Yahara, I. & Tanaka, K. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 22, 3557–3567 (2003)
Isono, E. et al. The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol. Biol. Cell 18, 569–580 (2007)
Kusmierczyk, A. R., Kunjappu, M. J., Funakoshi, M. & Hochstrasser, M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nature Struct. Mol. Biol. 15, 237–244 (2008)
Kusmierczyk, A. R. & Hochstrasser, M. Some assembly required: dedicated chaperones in eukaryotic proteasome biogenesis. Biol. Chem. 389, 1143–1151 (2008)
Xie, Y. & Varshavsky, A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl Acad. Sci. USA 98, 3056–3061 (2001)
Kleijnen, M. F. et al. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nature Struct. Mol. Biol. 14, 1180–1188 (2007)
Forster, A., Masters, E. I., Whitby, F. G., Robinson, H. & Hill, C. P. The 1.9 Å structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol. Cell 18, 589–599 (2005)
Gillette, T. G., Kumar, B., Thompson, D., Slaughter, C. A. & Demartino, G. N. Differential roles of the C-termini of AAA subunits of PA700 (19S regulator) in asymmetric assembly and activation of the 26S proteasome. J. Biol. Chem. 283, 31813–31822 (2008)
Smith, D. M. et al. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol. Cell 27, 731–744 (2007)
Ammelburg, M., Frickey, T. & Lupas, A. N. Classification of AAA+ proteins. J. Struct. Biol. 156, 2–11 (2006)
Nakamura, Y. et al. Structural basis for the recognition between the regulatory particles Nas6 and Rpt3 of the yeast 26S proteasome. Biochem. Biophys. Res. Commun. 359, 503–509 (2007)
Nakamura, Y. et al. Structure of the oncoprotein gankyrin in complex with S6 ATPase of the 26S proteasome. Structure 15, 179–189 (2007)
Zhang, F. et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii . Mol. Cell (in the press)
Groll, M. et al. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 386, 463–471 (1997)
Gorbea, C., Taillandier, D. & Rechsteiner, M. Mapping subunit contacts in the regulatory complex of the 26S proteasome. S2 and S5b form a tetramer with ATPase subunits S4 and S7. J. Biol. Chem. 275, 875–882 (2000)
Le Tallec, B., Barrault, M. B., Guerois, R., Carre, T. & Peyroche, A. Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome. Mol. Cell 33, 389–399 (2009)
Ellis, R. J. Molecular chaperones: assisting assembly in addition to folding. Trends Biochem. Sci. 31, 395–401 (2006)
Hirano, Y. et al. Dissecting beta-ring assembly pathway of the mammalian 20S proteasome. EMBO J. 27, 2204–2213 (2008)
Effantin, G., Rosenzweig, R., Glickman, M. & Steven, A. C. Electron microscopic evidence in support of α-solenoid models of proteasomal subunits Rpn1 and Rpn2. J. Mol. Biol. 386, 1204–1211 (2009)
Rosenzweig, R., Osmulski, P. A., Gaczynska, M. & Glickman, M. H. The central unit within the 19S regulatory particle of the proteasome. Nature Struct. Mol. Biol. 15, 573–580 (2008)
Acknowledgements
We thank C. Mann for Rpt1 (also known as Cim5) and Rpt6 (Cim3) antibodies, K. Kubota for preparing HeLa lysates and especially L. Huang for the human Rpn11-HTBH-expressing cell line. We also thank J. Hanna, M. Schmidt and members of the Finley laboratory, in particular S. Elsasser, for critically reading the manuscript. This work was supported by the NIH (GM043601 to D.F. and GM67945 to S.P.G.), a NIH NRSA postdoctoral fellowship (5F32GM75737-2 to S.P.) and an EMBO long-term fellowship (to J.R.).
Author Contributions J.R., Y.H. and F.E.M. performed the experiments. W.H., F.E.M. and S.P.G. performed mass spectrometry. G.T. performed modelling. F.Z. and Y.S. contributed to modelling. B.-H.L. purified human proteasomes. J.R., S.P. and D.F. interpreted data and developed the model. J.R. and D.F. planned the studies and wrote the manuscript. All authors commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-9 with Legends, Supplementary Tables 1-4, Supplementary Methods and Supplementary References. (PDF 3849 kb)
Rights and permissions
About this article
Cite this article
Roelofs, J., Park, S., Haas, W. et al. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature 459, 861–865 (2009). https://doi.org/10.1038/nature08063
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08063
This article is cited by
-
Gankyrin inhibits ferroptosis through the p53/SLC7A11/GPX4 axis in triple-negative breast cancer cells
Scientific Reports (2023)
-
Tagging the proteasome active site β5 causes tag specific phenotypes in yeast
Scientific Reports (2020)
-
Co-translational assembly of proteasome subunits in NOT1-containing assemblysomes
Nature Structural & Molecular Biology (2019)
-
Regulation of proteasome assembly and activity in health and disease
Nature Reviews Molecular Cell Biology (2018)
-
Structural insights on the dynamics of proteasome formation
Biophysical Reviews (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.