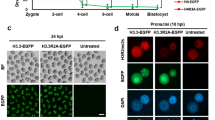
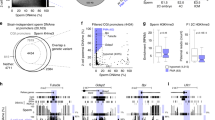
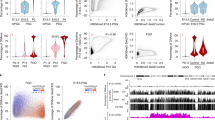
Abstract
The life cycle of mammals begins when a sperm enters an egg. Immediately after fertilization, both the maternal and paternal genomes undergo dramatic reprogramming to prepare for the transition from germ cell to somatic cell transcription programs1. One of the molecular events that takes place during this transition is the demethylation of the paternal genome2,3. Despite extensive efforts, the factors responsible for paternal DNA demethylation have not been identified4. To search for such factors, we developed a live cell imaging system that allows us to monitor the paternal DNA methylation state in zygotes. Through short-interfering-RNA-mediated knockdown in mouse zygotes, we identified Elp3 (also called KAT9), a component of the elongator complex5, to be important for paternal DNA demethylation. We demonstrate that knockdown of Elp3 impairs paternal DNA demethylation as indicated by reporter binding, immunostaining and bisulphite sequencing. Similar results were also obtained when other elongator components, Elp1 and Elp4, were knocked down. Importantly, injection of messenger RNA encoding the Elp3 radical SAM domain mutant, but not the HAT domain mutant, into MII oocytes before fertilization also impaired paternal DNA demethylation, indicating that the SAM radical domain is involved in the demethylation process. Our study not only establishes a critical role for the elongator complex in zygotic paternal genome demethylation, but also indicates that the demethylation process may be mediated through a reaction that requires an intact radical SAM domain.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
28 January 2010
In the paragraph beginning 'To provide direct evidence...', the reference to Fig. 3c was corrected to Fig. 2c on 28 January 2010.
References
Reik, W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447, 425–432 (2007)
Mayer, W., Niveleau, A., Walter, J., Fundele, R. & Haaf, T. Demethylation of the zygotic paternal genome. Nature 403, 501–502 (2000)
Oswald, J. et al. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 10, 475–478 (2000)
Ooi, S. K. & Bestor, T. H. The colorful history of active DNA demethylation. Cell 133, 1145–1148 (2008)
Svejstrup, J. Q. Elongator complex: how many roles does it play? Curr. Opin. Cell Biol. 19, 331–336 (2007)
Howell, C. Y. et al. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 104, 829–838 (2001)
Hajkova, P. et al. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 117, 15–23 (2002)
Bhattacharya, S. K., Ramchandani, S., Cervoni, N. & Szyf, M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature 397, 579–583 (1999)
Santos, F., Hendrich, B., Reik, W. & Dean, W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 241, 172–182 (2002)
Choi, Y. et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis . Cell 110, 33–42 (2002)
Gong, Z. et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814 (2002)
Rai, K. et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135, 1201–1212 (2008)
Barreto, G. et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445, 671–675 (2007)
Ma, D. K. et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323, 1074–1077 (2009)
Engel, N. et al. Conserved DNA methylation in Gadd45a-/- mice. Epigenetics 4, 98–99 (2009)
Jin, S. G., Guo, C. & Pfeifer, G. P. GADD45A does not promote DNA demethylation. PLoS Genet. 4, e1000013 (2008)
Allen, M. D. et al. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. EMBO J. 25, 4503–4512 (2006)
Jorgensen, H. F., Adie, K., Chaubert, P. & Bird, A. P. Engineering a high-affinity methyl-CpG-binding protein. Nucleic Acids Res. 34, e96 (2006)
Yamagata, K., Suetsugu, R. & Wakayama, T. Long-term, six-dimensional live-cell imaging for the mouse preimplantation embryo that does not affect full-term development. J. Reprod. Dev. 55, 343–350 (2009)
Torres-Padilla, M. E., Bannister, A. J., Hurd, P. J., Kouzarides, T. & Zernicka-Goetz, M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 50, 455–461 (2006)
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009)
Kim, S. H. et al. Differential DNA methylation reprogramming of various repetitive sequences in mouse preimplantation embryos. Biochem. Biophys. Res. Commun. 324, 58–63 (2004)
Lane, N. et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35, 88–93 (2003)
Wittschieben, B. O. et al. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4, 123–128 (1999)
Hawkes, N. A. et al. Purification and characterization of the human elongator complex. J. Biol. Chem. 277, 3047–3052 (2002)
Wang, S. C. & Frey, P. A. S-adenosylmethionine as an oxidant: the radical SAM superfamily. Trends Biochem. Sci. 32, 101–110 (2007)
Paraskevopoulou, C., Fairhurst, S. A., Lowe, D. J., Brick, P. & Onesti, S. The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol. Microbiol. 59, 795–806 (2006)
Tagami, H., Ray-Gallet, D., Almouzni, G. & Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 (2004)
Opavsky, R. et al. CpG island methylation in a mouse model of lymphoma is driven by the genetic configuration of tumor cells. PLoS Genet. 3, e167 (2007)
Tremblay, K. D., Duran, K. L. & Bartolomei, M. S. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell. Biol. 17, 4322–4329 (1997)
Jackson-Grusby, L. et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nature Genet. 27, 31–39 (2001)
Acknowledgements
We thank H. Song for providing the Gadd45b null mice, and K. Aoki and M. Matsuda for helping with image quantification. We are grateful to S. Wu for critical reading of the manuscript. Y.Z. is an investigator of the Howard Hughes Medical Institute.
Author Contributions Y.Z. conceived the project. Y.Z. and Y.O. designed the experiments and prepared the manuscript. Y.O. performed the majority of the experiments. K.Y. and K.H. helped with some of the experiments. T.W. provided the instruments and reagent for Y.O.’s technical training.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures S1-S8 with Legends and Supplementary Tables S1-S3. (PDF 1753 kb)
Rights and permissions
About this article
Cite this article
Okada, Y., Yamagata, K., Hong, K. et al. A role for the elongator complex in zygotic paternal genome demethylation. Nature 463, 554–558 (2010). https://doi.org/10.1038/nature08732
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08732
This article is cited by
-
Recurrent RNA edits in human preimplantation potentially enhance maternal mRNA clearance
Communications Biology (2022)
-
The Elongator subunit Elp3 is a non-canonical tRNA acetyltransferase
Nature Communications (2019)
-
Structural insights into the function of Elongator
Cellular and Molecular Life Sciences (2018)
-
The Arabidopsis ELP3/ELO3 and ELP4/ELO1 genes enhance disease resistance in Fragaria vesca L.
BMC Plant Biology (2017)
-
Elongator Protein 3 (Elp3) stabilizes Snail1 and regulates neural crest migration in Xenopus
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.