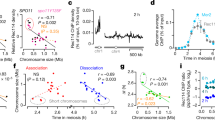
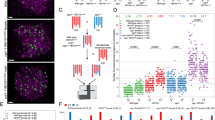
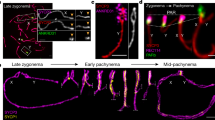
Abstract
Meiotic crossover (CO) recombination establishes physical linkages between homologous chromosomes that are required for their proper segregation into developing gametes, and promotes genetic diversity by shuffling genetic material between parental chromosomes. COs require the formation of double strand breaks (DSBs) to create the substrate for strand exchange. DSBs occur in small intervals called hotspots1,2,3 and significant variation in hotspot usage exists between and among individuals4. This variation is thought to reflect differences in sequence identity and chromatin structure, DNA topology and/ or chromosome domain organization1,5,6,7,8,9. Chromosomes show different frequencies of nondisjunction (NDJ)10, reflecting inherent differences in meiotic crossover control, yet the underlying basis of these differences remains elusive. Here we show that a novel chromatin factor, X non-disjunction factor 1 (xnd-1), is responsible for the global distribution of COs in C. elegans. xnd-1 is also required for formation of double-strand breaks (DSBs) on the X, but surprisingly XND-1 protein is autosomally enriched. We show that xnd-1 functions independently of genes required for X chromosome-specific gene silencing, revealing a novel pathway that distinguishes the X from autosomes in the germ line, and further show that xnd-1 exerts its effects on COs, at least in part, by modulating levels of H2A lysine 5 acetylation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gerton, J. L. et al. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae . Proc. Natl Acad. Sci. USA 97, 11383–11390 (2000)
Buhler, C., Borde, V. & Lichten, M. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae . PLoS Biol. 5, e324 (2007)
Mancera, E., Bourgon, R., Brozzi, A., Huber, W. & Steinmetz, L. M. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454, 479–485 (2008)
Coop, G., Wen, X., Ober, C., Pritchard, J. K. & Przeworski, M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science 319, 1395–1398 (2008)
Robine, N. et al. Genome-wide redistribution of meiotic double-strand breaks in Saccharomyces cerevisiae . Mol. Cell. Biol 27, 1868–1880 (2007)
Mieczkowski, P. A., Dominska, M., Buck, M. J., Lieb, J. D. & Petes, T. D. Loss of a histone deacetylase dramatically alters the genomic distribution of Spo11p-catalyzed DNA breaks in Saccharomyces cerevisiae . DNA Repair (Amst.) 104, 3955–3960 (2007)
Buard, J., Barthes, P., Grey, C. & de Massy, B. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 28, 2616–2624 (2009)
Borde, V. et al. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 28, 99–111 (2008)
Mets, D. G. & Meyer, B. J. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139, 73–86 (2009)
Hassold, T., Hall, H. & Hunt, P. The origin of human aneuploidy: where we have been, where we are going. Hum. Mol. Genet. 16, R203–R208 (2007)
Barnes, T. M., Kohara, Y., Coulson, A. & Hekimi, S. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans . Genetics 141, 159–179 (1995)
Rockman, M. V. & Kruglyak, L. Recombinational landscape and population genomics of Caenorhabditis elegans . PLoS Genet. 5, e1000419 (2009)
Kelly, W. G. et al. X-chromosome silencing in the germline of C. elegans . Development 129, 479–492 (2002)
Hodgkin, J., Horvitz, H. R. & Brenner, S. Nondisjunction mutants of the nematode Caenorhabditis elegans . Genetics 91, 67–94 (1979)
Keeney, S., Giroux, C. N. & Kleckner, N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997)
Dernburg, A. F. et al. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94, 387–398 (1998)
Petty, E. L., Collette, K. S., Cohen, A. J., Snyder, M. J. & Csankovszki, G. Restricting dosage compensation complex binding to the X chromosomes by H2A.Z/HTZ-1. PLoS Genet. 5, e1000699 (2009)
Smolikov, S., Schild-Prufert, K. & Colaiacovo, M. P. CRA-1 uncovers a double-strand break-dependent pathway promoting the assembly of central region proteins on chromosome axes during C. elegans meiosis. PLoS Genet. 4, e1000088 (2008)
Fong, Y., Bender, L., Wang, W. & Strome, S. Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans . Science 296, 2235–2238 (2002)
Kelly, W. G., Xu, S., Montgomery, M. K. & Fire, A. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics 146, 227–238 (1997)
Ikura, T. et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102, 463–473 (2000)
Ceol, C. J. & Horvitz, H. R. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev. Cell 6, 563–576 (2004)
Zetka, M. C. & Rose, A. M. Mutant rec-1 eliminates the meiotic pattern of crossing over in Caenorhabditis elegans . Genetics 141, 1339–1349 (1995)
Baudat, F. et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840 (2010)
Myers, S. et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327, 876–879 (2010)
Parvanov, E. D., Petkov, P. M. & Paigen, K. Prdm9 controls activation of mammalian recombination hotspots. Science 327, 835 (2010)
Myers, S., Freeman, C., Auton, A., Donnelly, P. & McVean, G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nature Genet. 40, 1124–1129 (2008)
Hayashi, K., Yoshida, K. & Matsui, Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438, 374–378 (2005)
Brenner, S. The genetics of Caenorhabditis elegans . Genetics 77, 71–94 (1974)
Chan, A. & Meyer, B. J. Protocol 21: antibody staining of C . elegans gonads in Wormbook 〈http://www.wormbook.org/chapters/www_intromethodscellbiology/intromethodscellbiology.html#d0e2311〉 (2006)
Timmons, L. & Fire, A. Specific interference by ingested dsRNA. Nature 395, 854 (1998)
Colaiácovo, M. P. et al. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5, 463–474 (2003)
Phillips, C. M. et al. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123, 1051–1063 (2005)
Tsai, C. J. et al. Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev. 22, 194–211 (2008)
Lim, J. G., Stine, R. R. & Yanowitz, J. L. Domain-specific regulation of recombination in Caenorhabditis elegans in response to temperature, age and sex. Genetics 180, 715–726 (2008)
Davis, M. W., Hammarlund, M., Harrach, T., Hullett, S. O. & Jorgensen, E. M. Rapid single nucleotide polymorphism mapping in C. elegans. BMC . Genomics 6, 118 (2005)
Acknowledgements
We thank J. Bembenek, A. Bortvin, G. Deshpande, M. Halpern, V. Jantsch, A. MacQueen and D. Mets for comments on the manuscript; the Koshland lab and Baltimore Area Worm Club for discussions; M. Siddiqi for microscopy support. We are indebted to the Caenorhabditis Genetics Center and the C. elegans Knockout Consortium for worm stocks and Y. Kohara for cDNAs. This work was supported by the Carnegie Institution of Washington, NIH K01AG031296, and MWRI start-up funds to J.L.Y. D.L.B. was supported by a Canada Research Chair and a grant from NSERC.
Author information
Authors and Affiliations
Contributions
C.R.W. performed RNAi screen, SNP analysis, FISH and immunofluorescence. L.K. and D.L.B. identified xnd-1 as a HIM mutant, created co-suppression lines, and determined hatching rates and embryonic lethality. J.L.Y. prepared samples for immunofluorescence, isolated protein for antibody production and performed confocal microscopy. C.R.W. and J.L.Y. designed experiments, analysed data, and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-7 with legends Supplementary Tables 1-6, legends for Supplementary Movies 1-3 and additional references. (PDF 8507 kb)
Supplementary Movie 1
This movie shows a 3D reconstruction of confocal stocks of a wild type gonad revealing RAD-51 foci on the X chromosome indicating the formation of DSBs on the X - see Supplementary Information file for full legend. (MOV 5771 kb)
Supplementary Movie 2
This movie shows a 3D reconstruction of confocal stocks of a xnd-1 mutant gonad revealing the absence of RAD-51 foci on the X chromosome, revealing the lack of DSBs on a fraction of X chromosomes in the mutant - see Supplementary Information file for full legend. (MOV 4182 kb)
Supplementary Movie 3
This movie shows a 3D reconstruction of confocal stacks from wild type gonads showing the enrichment of XND-1 protein on autosomes - see Supplementary Information file for full legend. (MOV 7313 kb)
Rights and permissions
About this article
Cite this article
Wagner, C., Kuervers, L., Baillie, D. et al. xnd-1 regulates the global recombination landscape in Caenorhabditis elegans. Nature 467, 839–843 (2010). https://doi.org/10.1038/nature09429
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09429
This article is cited by
-
The genome loading model for the origin and maintenance of sex in eukaryotes
Biologia Futura (2022)
-
Poly(ADP-ribose) glycohydrolase coordinates meiotic DNA double-strand break induction and repair independent of its catalytic activity
Nature Communications (2020)
-
Leagues of their own: sexually dimorphic features of meiotic prophase I
Chromosoma (2019)
-
Epigenetic regulation of genomic integrity
Chromosoma (2012)
-
Meiotic crossover: what controls the breaks?
Asian Journal of Andrology (2011)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.