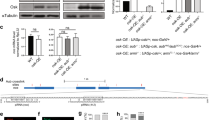
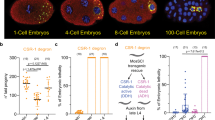
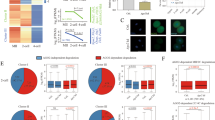
Abstract
Piwi-associated RNAs (piRNAs), a specific class of 24- to 30-nucleotide-long RNAs produced by the Piwi-type of Argonaute proteins, have a specific germline function in repressing transposable elements. This repression is thought to involve heterochromatin formation and transcriptional and post-transcriptional silencing1,2,3,4,5,6. The piRNA pathway has other essential functions in germline stem cell maintenance7 and in maintaining germline DNA integrity8,9,10. Here we uncover an unexpected function of the piRNA pathway in the decay of maternal messenger RNAs and in translational repression in the early embryo. A subset of maternal mRNAs is degraded in the embryo at the maternal-to-zygotic transition. In Drosophila, maternal mRNA degradation depends on the RNA-binding protein Smaug and the deadenylase CCR411,12,13, as well as the zygotic expression of a microRNA cluster14. Using mRNA encoding the embryonic posterior morphogen Nanos (Nos) as a paradigm to study maternal mRNA decay, we found that CCR4-mediated deadenylation of nos depends on components of the piRNA pathway including piRNAs complementary to a specific region in the nos 3′ untranslated region. Reduced deadenylation when piRNA-induced regulation is impaired correlates with nos mRNA stabilization and translational derepression in the embryo, resulting in head development defects. Aubergine, one of the Argonaute proteins in the piRNA pathway, is present in a complex with Smaug, CCR4, nos mRNA and piRNAs that target the nos 3′ untranslated region, in the bulk of the embryo. We propose that piRNAs and their associated proteins act together with Smaug to recruit the CCR4 deadenylation complex to specific mRNAs, thus promoting their decay. Because the piRNAs involved in this regulation are produced from transposable elements, this identifies a direct developmental function for transposable elements in the regulation of gene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Saito, K. et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20, 2214–2222 (2006)
Vagin, V. V. et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324 (2006)
Brennecke, J. et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 (2007)
Yin, H. & Lin, H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450, 304–308 (2007)
Aravin, A. A. et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799 (2008)
Lim, A. K., Tao, L. & Kai, T. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J. Cell Biol. 186, 333–342 (2009)
Cox, D. N. et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715–3727 (1998)
Chen, Y., Pane, A. & Schupbach, T. cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr. Biol. 17, 637–642 (2007)
Klattenhoff, C. et al. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 12, 45–55 (2007)
Pane, A., Wehr, K. & Schupbach, T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell 12, 851–862 (2007)
Semotok, J. L. et al. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr. Biol. 15, 284–294 (2005)
Tadros, W. et al. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev. Cell 12, 143–155 (2007)
Zaessinger, S., Busseau, I. & Simonelig, M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development 133, 4573–4583 (2006)
Bushati, N., Stark, A., Brennecke, J. & Cohen, S. M. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 18, 501–506 (2008)
Gavis, E. R. & Lehmann, R. Translational regulation of nanos by RNA localization. Nature 369, 315–318 (1994)
Dahanukar, A. & Wharton, R. P. The Nanos gradient in Drosophila embryos is generated by translational regulation. Genes Dev. 10, 2610–2621 (1996)
Dahanukar, A., Walker, J. A. & Wharton, R. P. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol. Cell 4, 209–218 (1999)
Giraldez, A. J. et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79 (2006)
Chung, W. J., Okamura, K., Martin, R. & Lai, E. C. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. 18, 795–802 (2008)
Brennecke, J. et al. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322, 1387–1392 (2008)
Gunawardane, L. S. et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315, 1587–1590 (2007)
Li, C. et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137, 509–521 (2009)
Cook, H. A., Koppetsch, B. S., Wu, J. & Theurkauf, W. E. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 116, 817–829 (2004)
Malone, C. D. et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137, 522–535 (2009)
Harris, A. N. & Macdonald, P. M. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 128, 2823–2832 (2001)
Megosh, H. B., Cox, D. N., Campbell, C. & Lin, H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr. Biol. 16, 1884–1894 (2006)
Lehmann, R. & Nusslein-Volhard, C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development 112, 679–691 (1991)
Leaman, D. et al. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell 121, 1097–1108 (2005)
Saito, K. et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461, 1296–1299 (2009)
Robine, N. et al. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr. Biol. 19, 2066–2076 (2009)
Schupbach, T. & Wieschaus, E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129, 1119–1136 (1991)
Gillespie, D. E. & Berg, C. A. homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes Dev. 9, 2495–2508 (1995)
Chou, T. B. & Perrimon, N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144, 1673–1679 (1996)
Wang, C., Dickinson, L. K. & Lehmann, R. Genetics of nanos localization in Drosophila. Dev. Dyn. 199, 103–115 (1994)
Rorth, P. Gal4 in the Drosophila female germline. Mech. Dev. 78, 113–118 (1998)
Juge, F., Zaessinger, S., Temme, C., Wahle, E. & Simonelig, M. Control of poly(A) polymerase level is essential to cytoplasmic polyadenylation and early development in Drosophila. EMBO J. 21, 6603–6613 (2002)
Pelisson, A., Sarot, E., Payen-Groschene, G. & Bucheton, A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J. Virol. 81, 1951–1960 (2007)
Benoit, B. et al. An essential cytoplasmic function for the nuclear poly(A) binding protein, PABP2, in poly(A) tail length control and early development in Drosophila. Dev. Cell 9, 511–522 (2005)
Benoit, B. et al. The Drosophila poly(A)-binding protein II is ubiquitous throughout Drosophila development and has the same function in mRNA polyadenylation as its bovine homolog in vitro. Nucleic Acids Res. 27, 3771–3778 (1999)
Temme, C., Zaessinger, S., Meyer, S., Simonelig, M. & Wahle, E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 23, 2862–2871 (2004)
Acknowledgements
We are grateful to A. Nakamura, M. Siomi, H. Siomi, C. Smibert, H. Lin, P. Macdonald, T. Schüpbach, W. Theurkauf, R. Wharton and P. Zamore, for their gifts of antibodies or Drosophila stocks. We thank M. Benkirane for the gift of 2′-O-methyl anti-miR129. This work was supported by the Centre National de la Recherche Scientifique UPR1142, the Agence Nationale de la Recherche (ANR Blanche ANR-06-BLAN-0343), the Fondation pour la Recherche Médicale (FRM, Equipe FRM 2007) and the Association pour la Recherche sur le Cancer (ARC Libre 2009) to M.S. and by the National Institutes of Health (NIH R01-GM083300) to E.C.L. C.R., A.C.M. and B.F. held salaries from ANR Blanche.
Author information
Authors and Affiliations
Contributions
C.R. and C.P. designed and performed the experiments, analysed the data and contributed equally to the study. A.-C.M. contributed to the generation of DNA constructs, B.F. contributed to poly(A) test assays in Fig. 1. A.B., N.R. and E.C.L. performed the bioinformatic analyses. A.P. performed northern blots. M.S. designed the study, analysed data and wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion, Supplementary Figures 1-12 with legends and additional references. (PDF 4507 kb)
Rights and permissions
About this article
Cite this article
Rouget, C., Papin, C., Boureux, A. et al. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467, 1128–1132 (2010). https://doi.org/10.1038/nature09465
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09465
This article is cited by
-
Critical appraisal of the piRNA-PIWI axis in cancer and cancer stem cells
Biomarker Research (2024)
-
The burgeoning importance of PIWI-interacting RNAs in cancer progression
Science China Life Sciences (2024)
-
Themes and variations on piRNA-guided transposon control
Mobile DNA (2023)
-
The non-redundant functions of PIWI family proteins in gametogenesis in golden hamsters
Nature Communications (2023)
-
Emerging roles and functional mechanisms of PIWI-interacting RNAs
Nature Reviews Molecular Cell Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.