Abstract
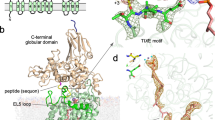
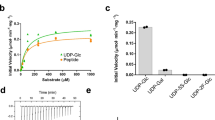
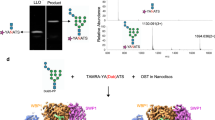
Asparagine-linked glycosylation is a post-translational modification of proteins containing the conserved sequence motif Asn-X-Ser/Thr. The attachment of oligosaccharides is implicated in diverse processes such as protein folding and quality control, organism development or host–pathogen interactions. The reaction is catalysed by oligosaccharyltransferase (OST), a membrane protein complex located in the endoplasmic reticulum. The central, catalytic enzyme of OST is the STT3 subunit, which has homologues in bacteria and archaea. Here we report the X-ray structure of a bacterial OST, the PglB protein of Campylobacter lari, in complex with an acceptor peptide. The structure defines the fold of STT3 proteins and provides insight into glycosylation sequon recognition and amide nitrogen activation, both of which are prerequisites for the formation of the N-glycosidic linkage. We also identified and validated catalytically important, acidic amino acid residues. Our results provide the molecular basis for understanding the mechanism of N-linked glycosylation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Apweiler, R., Hermjakob, H. & Sharon, N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 (1999)
Zielinska, D. F., Gnad, F., Wisniewski, J. R. & Mann, M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 (2010)
Helenius, A. & Aebi, M. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 (2004)
Kornfeld, R. & Kornfeld, S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54, 631–664 (1985)
Varki, A. Biological roles of oligosaccharides—all of the theories are correct. Glycobiology 3, 97–130 (1993)
Kelleher, D. J. & Gilmore, R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R–62R (2006)
Petrescu, A. J., Milac, A. L., Petrescu, S. M., Dwek, R. A. & Wormald, M. R. Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology 14, 103–114 (2004)
Breton, C. & Imberty, A. Structure/function studies of glycosyltransferases. Curr. Opin. Struct. Biol. 9, 563–571 (1999)
Charnock, S. J. & Davies, G. J. Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry 38, 6380–6385 (1999)
Ünligil, U. M. & Rini, J. M. Glycosyltransferase structure and mechanism. Curr. Opin. Struct. Biol. 10, 510–517 (2000)
Lairson, L. L., Henrissat, B., Davies, G. J. & Withers, S. G. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 (2008)
Lazarus, M. B., Nam, Y., Jiang, J., Sliz, P. & Walker, S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469, 564–567 (2011)
Chen, W. & Helenius, A. Role of ribosome and translocon complex during folding of influenza hemagglutinin in the endoplasmic reticulum of living cells. Mol. Biol. Cell 11, 765–772 (2000)
Silberstein, S. & Gilmore, R. Biochemistry, molecular biology, and genetics of the oligosaccharyltransferase. FASEB J. 10, 849–858 (1996)
Whitley, P., Nilsson, I. & von Heijne, G. A nascent secretory protein may traverse the ribosome endoplasmic reticulum translocase complex as an extended chain. J. Biol. Chem. 271, 6241–6244 (1996)
Ruiz-Canada, C., Kelleher, D. J. & Gilmore, R. Cotranslational and Posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136, 272–283 (2009)
Zufferey, R. et al. Stt3, a highly conserved protein required for yeast oligosaccharyl transferase activity in vivo. EMBO J. 14, 4949–4960 (1995)
Wilson, C. M. & High, S. Ribophorin I acts as a substrate-specific facilitator of N-glycosylation. J. Cell Sci. 120, 648–657 (2007)
Schulz, B. L. et al. Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency. Proc. Natl Acad. Sci. USA 106, 11061–11066 (2009)
Mescher, M. F. & Strominger, J. L. Purification and characterization of a prokaryotic glycoprotein from cell-envelope of Halobacterium salinarium. J. Biol. Chem. 251, 2005–2014 (1976)
Szymanski, C. M., Yao, R. J., Ewing, C. P., Trust, T. J. & Guerry, P. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32, 1022–1030 (1999)
Izquierdo, L. et al. Distinct donor and acceptor specificities of Trypanosoma brucei oligosaccharyltransferases. EMBO J. 28, 2650–2661 (2009)
Nasab, F. P., Schulz, B. L., Gamarro, F., Parodi, A. J. & Aebi, M. All in one: Leishmania major STT3 proteins substitute for the whole oligosaccharyltransferase complex in Saccharomyces cerevisiae. Mol. Biol. Cell 19, 3758–3768 (2008)
Szymanski, C. M. & Wren, B. W. Protein glycosylation in bacterial mucosal pathogens. Nature Rev. Microbiol. 3, 225–237 (2005)
Wacker, M. et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298, 1790–1793 (2002)
Abu-Qarn, M. & Eichler, J. Protein N-glycosylation in Archaea: defining Haloferax volcanii genes involved in S-layer glycoprotein glycosylation. Mol. Microbiol. 61, 511–525 (2006)
Li, L. et al. Overexpression and topology of bacterial oligosaccharyltransferase PglB. Biochem. Biophys. Res. Commun. 394, 1069–1074 (2010)
Schwarz, F. et al. Relaxed acceptor site specificity of bacterial oligosaccharyltransferase in vivo. Glycobiology 21, 45–54 (2011)
Chen, M. M., Glover, K. J. & Imperiali, B. From peptide to protein: Comparative analysis of the substrate specificity of N-linked glycosylation in C. jejuni. Biochemistry 46, 5579–5585 (2007)
Igura, M. et al. Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J. 27, 234–243 (2008)
Maita, N., Nyirenda, J., Igura, M., Kamishikiryo, J. & Kohda, D. Comparative structural biology of eubacterial and archaeal oligosaccharyltransferases. J. Biol. Chem. 285, 4941–4950 (2010)
Collaborative Computational Project, 4. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Kowarik, M. et al. N-linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science 314, 1148–1150 (2006)
Bause, E. & Legler, G. The role of the hydroxy amino acid in the triplet sequence Asn-Xaa-Thr(Ser) for the N-glycosylation step during glycoprotein biosynthesis. Biochem. J. 195, 639–644 (1981)
Kowarik, M. et al. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 25, 1957–1966 (2006)
Bause, E. Model studies on N-glycosylation of proteins. Biochem. Soc. Trans. 12, 514–517 (1984)
Breuer, W., Klein, R. A., Hardt, B., Bartoschek, A. & Bause, E. Oligosaccharyltransferase is highly specific for the hydroxy amino acid in Asn-Xaa-Thr/Ser. FEBS Lett. 501, 106–110 (2001)
Valliere-Douglass, J. F. et al. Asparagine-linked oligosaccharides present on a non-consensus amino acid sequence in the CH1 domain of human antibodies. J. Biol. Chem. 284, 32493–32506 (2009)
Imperiali, B. & Rickert, K. W. Conformational implications of asparagine-linked glycosylation. Proc. Natl Acad. Sci. USA 92, 97–101 (1995)
Sharma, C., Lehle, L. & Tanner, W. N-glycosylation of yeast proteins—characterization of the solubilized oligosaccharyl transferase. Eur. J. Biochem. 116, 101–108 (1981)
Liu, J. & Mushegian, A. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 12, 1418–1431 (2003)
Maeda, Y. et al. PIG-M transfers the first mannose to glycosylphosphatidylinositol on the lumenal side of the ER. EMBO J. 20, 250–261 (2001)
Imperiali, B., Shannon, K. L. & Rickert, K. W. Role of peptide conformation in asparagine-linked glycosylation. J. Am. Chem. Soc. 114, 7942–7944 (1992)
Wiberg, K. B. & Breneman, C. M. Resonance interactions in acyclic systems. 3. Formamide internal rotation revisited. Charge and energy redistribution along the C-N bond rotational pathway. J. Am. Chem. Soc. 114, 831–840 (1992)
Cleland, W. W. & Kreevoy, M. M. Low-barrier hydrogen bonds and enzymatic catalysis. Science 264, 1887–1890 (1994)
Li, H., Chavan, M., Schindelin, H., Lennarz, W. J. & Li, H. L. Structure of the oligosaccharyl transferase complex at 12 Å resolution. Structure 16, 432–440 (2008)
Lizak, C., Fan, Y. Y., Weber, T. C. & Aebi, M. N-linked glycosylation of antibody fragments in Escherichia coli. Bioconjug. Chem. 22, 488–496 (2011)
Lefebre, M. D. & Valvano, M. A. Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 68, 5956–5964 (2002)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Strong, M. et al. Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Acknowledgements
We thank the beamline staff at the Swiss Light Source for assistance with data collection, S. Fleurkens and M. Bucher for technical assistance, and D. Arigoni for discussions. E. coli SCM6 was provided by C. Marolda and M. Valvano. C.L. was affiliated with the Life Science Zurich Graduate School. This research was supported by the NCCR Structural Biology Zurich (grant to K.P.L.) and Swiss National Science Foundation grants to M.A. (SNF 31003A-127098/1) and K.P.L. (SNF 31003A-131075/1).
Author information
Authors and Affiliations
Contributions
C.L., M.A. and K.P.L. designed the experiments. C.L., S.G. and S.N. performed the experiments; K.P.L. performed crystallographic calculations and model building; C.L., M.A. and K.P.L. analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
ETH Zurich has filed a patent application for the use of the PglB structure for biotechnological applications.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-8 with legends, Supplementary Table 1 and additional references. (PDF 3936 kb)
Rights and permissions
About this article
Cite this article
Lizak, C., Gerber, S., Numao, S. et al. X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–355 (2011). https://doi.org/10.1038/nature10151
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10151
This article is cited by
-
Progress towards a glycoconjugate vaccine against Group A Streptococcus
npj Vaccines (2023)
-
Molecular basis for bacterial N-glycosylation by a soluble HMW1C-like N-glycosyltransferase
Nature Communications (2023)
-
Endoplasmic reticulum-quality control pathway and endoplasmic reticulum-associated degradation mechanism regulate the N-glycoproteins and N-glycan structures in the diatom Phaeodactylum tricornutum
Microbial Cell Factories (2022)
-
Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase
Nature Communications (2022)
-
Glycosyltransferases as targets for therapeutic intervention in cancer and inflammation: molecular modeling insights
Chemical Papers (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.