Abstract
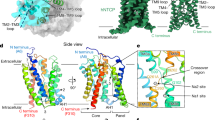
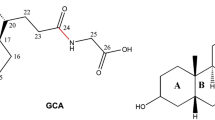
High cholesterol levels greatly increase the risk of cardiovascular disease. About 50 per cent of cholesterol is eliminated from the body by its conversion into bile acids. However, bile acids released from the bile duct are constantly recycled, being reabsorbed in the intestine by the apical sodium-dependent bile acid transporter (ASBT, also known as SLC10A2). It has been shown in animal models that plasma cholesterol levels are considerably lowered by specific inhibitors of ASBT1,2, and ASBT is thus a target for hypercholesterolaemia drugs. Here we report the crystal structure of a bacterial homologue of ASBT from Neisseria meningitidis (ASBTNM) at 2.2 Å. ASBTNM contains two inverted structural repeats of five transmembrane helices. A core domain of six helices harbours two sodium ions, and the remaining four helices pack in a row to form a flat, ‘panel’-like domain. Overall, the architecture of the protein is remarkably similar to the sodium/proton antiporter NhaA3, despite having no detectable sequence homology. The ASBTNM structure was captured with the substrate taurocholate present, bound between the core and panel domains in a large, inward-facing, hydrophobic cavity. Residues near this cavity have been shown to affect the binding of specific inhibitors of human ASBT4. The position of the taurocholate molecule, together with the molecular architecture, suggests the rudiments of a possible transport mechanism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lewis, M. C., Brieaddy, L. E. & Root, C. Effects of 2164U90 on ileal bile acid absorption and serum cholesterol in rats and mice. J. Lipid Res. 36, 1098–1105 (1995)
Bhat, B. G. et al. Inhibition of ileal bile acid transport and reduced atherosclerosis in apoE−/− mice by SC-435. J. Lipid Res. 44, 1614–1621 (2003)
Hunte, C. et al. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 435, 1197–1202 (2005)
Hallén, S., Bjorquist, A., Ostlund-Lindqvist, A. M. & Sachs, G. Identification of a region of the ileal-type sodium/bile acid cotransporter interacting with a competitive bile acid transport inhibitor. Biochemistry 41, 14916–14924 (2002)
Hagenbuch, B. & Dawson, P. The sodium bile salt cotransport family SLC10. Pflugers Arch. 447, 566–570 (2004)
Wong, M. H., Oelkers, P., Craddock, A. L. & Dawson, P. A. Expression cloning and characterization of the hamster ileal sodium-dependent bile acid transporter. J. Biol. Chem. 269, 1340–1347 (1994)
Weinman, S. A., Carruth, M. W. & Dawson, P. A. Bile acid uptake via the human apical sodium-bile acid cotransporter is electrogenic. J. Biol. Chem. 273, 34691–34695 (1998)
Oelkers, P., Kirby, L. C., Heubi, J. E. & Dawson, P. A. Primary bile acid malabsorption caused by mutations in the ileal sodium-dependent bile acid transporter gene (SLC10A2). J. Clin. Invest. 99, 1880–1887 (1997)
Kramer, W. & Wess, G. Bile acid transport systems as pharmaceutical targets. Eur. J. Clin. Invest. 26, 715–732 (1996)
Drew, D., Lerch, M., Kunji, E., Slotboom, D. J. & de Gier, J. W. Optimization of membrane protein overexpression and purification using GFP fusions. Nature Methods 3, 303–313 (2006)
Sonoda, Y. et al. Benchmarking membrane protein detergent stability for improving throughput of high-resolution X-ray structures. Structure 19, 17–25 (2011)
Geyer, J., Wilke, T. & Petzinger, E. The solute carrier family SLC10: more than a family of bile acid transporters regarding function and phylogenetic relationships. Naunyn Schmiedebergs Arch. Pharmacol. 372, 413–431 (2006)
Sun, A. Q., Balasubramaniyan, N., Chen, H., Shahid, M. & Suchy, F. J. Identification of functionally relevant residues of the rat ileal apical sodium-dependent bile acid cotransporter. J. Biol. Chem. 281, 16410–16418 (2006)
Chignard, N. et al. Bile acid transport and regulating functions in the human biliary epithelium. Hepatology 33, 496–503 (2001)
Craddock, A. L. et al. Expression and transport properties of the human ileal and renal sodium-dependent bile acid transporter. Am. J. Physiol. 274, G157–G169 (1998)
Zheng, X., Ekins, S., Raufman, J. P. & Polli, J. E. Computational models for drug inhibition of the human apical sodium-dependent bile acid transporter. Mol. Pharm. 6, 1591–1603 (2009)
Banerjee, A. & Swaan, P. W. Membrane topology of human ASBT (SLC10A2) determined by dual label epitope insertion scanning mutagenesis. New evidence for seven transmembrane domains. Biochemistry 45, 943–953 (2006)
Hallén, S., Branden, M., Dawson, P. A. & Sachs, G. Membrane insertion scanning of the human ileal sodium/bile acid co-transporter. Biochemistry 38, 11379–11388 (1999)
Screpanti, E. & Hunte, C. Discontinuous membrane helices in transport proteins and their correlation with function. J. Struct. Biol. 159, 261–267 (2007)
Boudker, O. & Verdon, G. Structural perspectives on secondary active transporters. Trends Pharmacol. Sci. 31, 418–426 (2010)
Hagenbuch, B. & Meier, P. J. Sinusoidal (basolateral) bile salt uptake systems of hepatocytes. Semin. Liver Dis. 16, 129–136 (1996)
Zahner, D., Eckhardt, U. & Petzinger, E. Transport of taurocholate by mutants of negatively charged amino acids, cysteines, and threonines of the rat liver sodium-dependent taurocholate cotransporting polypeptide Ntcp. Eur. J. Biochem. 270, 1117–1127 (2003)
Olkhova, E., Hunte, C., Screpanti, E., Padan, E. & Michel, H. Multiconformation continuum electrostatics analysis of the NhaA Na+/H+ antiporter of Escherichia coli with functional implications. Proc. Natl Acad. Sci. USA 103, 2629–2634 (2006)
Hussainzada, N., Banerjee, A. & Swaan, P. W. Transmembrane domain VII of the human apical sodium-dependent bile acid transporter ASBT (SLC10A2) lines the substrate translocation pathway. Mol. Pharmacol. 70, 1565–1574 (2006)
Jardetzky, O. Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966)
Padan, E. The enlightening encounter between structure and function in the NhaA Na+-H+ antiporter. Trends Biochem. Sci. 33, 435–443 (2008)
Appel, M., Hizlan, D., Vinothkumar, K. R., Ziegler, C. & Kuhlbrandt, W. Conformations of NhaA, the Na+/H+ exchanger from Escherichia coli, in the pH-activated and ion-translocating states. J. Mol. Biol. 388, 659–672 (2009)
Forrest, L. R. et al. Mechanism for alternating access in neurotransmitter transporters. Proc. Natl Acad. Sci. USA 105, 10338–10343 (2008)
Shimamura, T. et al. Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science 328, 470–473 (2010)
Tzubery, T., Rimon, A. & Padan, E. Structure-based functional study reveals multiple roles of transmembrane segment IX and loop VIII–IX in NhaA Na+/H+ antiporter of Escherichia coli at physiological pH. J. Biol. Chem. 283, 15975–15987 (2008)
Drew, D. E., von Heijne, G., Nordlund, P. & de Gier, J. W. Green fluorescent protein as an indicator to monitor membrane protein overexpression in Escherichia coli. FEBS Lett. 507, 220–224 (2001)
Miroux, B. & Walker, J. E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 (1996)
Kawate, T. & Gouaux, E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006)
Drew, D. et al. GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae . Nature Protocols 3, 784–798 (2008)
Alexandrov, A. I., Mileni, M., Chien, E. Y., Hanson, M. A. & Stevens, R. C. Microscale fluorescent thermal stability assay for membrane proteins. Structure 16, 351–359 (2008)
Winter, G. Xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Collaborative Computational Project, Number 4 . The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Knight, S. D. RSPS version 4.0: a semi-interactive vector-search program for solving heavy-atom derivatives. Acta Crystallogr. D 56, 42–47 (2000)
de La Fortelle, E. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494 (1997)
Abrahams, J. P. & Leslie, A. G. W. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D 52, 30–42 (1996)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Jones, T. A. & Kjeldgaard, M. Electron-density map interpretation. Methods Enzymol. 277, 173–208 (1997)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Leslie, A. G. W. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 ESF-EACBM Newslett. Protein Crystallogr. 26, (1992)
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D 60, 2210–2221 (2004)
Winn, M. D., Isupov, M. N. & Murshudov, G. N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D 57, 122–133 (2001)
Harding, M. M. Metal-ligand geometry relevant to proteins and in proteins: sodium and potassium. Acta Crystallogr. D 58, 872–874 (2002)
Nayal, M. & Di Cera, E. Valence screening of water in protein crystals reveals potential Na+ binding sites. J. Mol. Biol. 256, 228–234 (1996)
Sanchez-Weatherby, J. et al. Improving diffraction by humidity control: a novel device compatible with X-ray beamlines. Acta Crystallogr. D 65, 1237–1246 (2009)
Kleywegt, G. J. & Jones, T. A. A super position. ESF/CCP4 Newslett. 31, 9–14 (1994)
Kleywegt, G. J. & Jones, T. A. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D 50, 178–185 (1994)
DeLano, W. L. PyMOL Molecular Viewer 〈http://www.pymol.org〉 (2002)
Potterton, L. et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D 60, 2288–2294 (2004)
Yamashita, A., Singh, S. K., Kawate, T., Jin, Y. & Gouaux, E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437, 215–223 (2005)
Acknowledgements
We are grateful to C. Lee and Y. Sekiguchi for assistance with cloning and expression screening of ASBTNM mutants, and to S. van de Graaf for donating fluorescently labelled bile acid, which was used in the initial functional characterization of ASBTNM. Data were collected at the European Synchrotron Radiation Facility, France, and the Diamond Light Source, UK, with assistance from beamline scientists; in particular, we would like to thank J. Sanchez-Weatherby for help with the HCl. We are also grateful to K. Beis and G. von Heijne for reading the manuscript. This work was funded by the Medical Research Council (MRC_G0900990(91997), to A.D.C. and D.D.), the European Union (EMeP grant LSHG-CT-2004-504601, to S.I.) and the Biotechnology and Biological Sciences Research Council (BB/G023425/1, to S.I.). Part of this work was also supported by a grant from the Targeted Proteins Research Program of MEXT, Japan, and the ERATO Iwata Human Receptor Crystallography Project, Japan Science and Technology Agency. The authors are grateful for the use of the Membrane Protein Laboratory funded by the Wellcome Trust (WT089809) at the Diamond Light Source. D.D. acknowledges personal support from The Royal Society through the University Research Fellow scheme.
Author information
Authors and Affiliations
Contributions
N.-J.H., S.I., A.D.C. and D.D. contributed to the design of the project. N.-J.H. and D.D. screened homologues, expressed and purified the protein, and carried out functional characterization. N.-J.H., S.I., A.D.C. and D.D. were involved in crystallographic experiments and analysis of data. A.C. and D.D. were responsible for overall project management and wrote the manuscript with assistance from N.-J.H. and S.I.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Tables 1-2, Supplementary Figures 1-10 with legends and additional references. (PDF 4000 kb)
Rights and permissions
About this article
Cite this article
Hu, NJ., Iwata, S., Cameron, A. et al. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature 478, 408–411 (2011). https://doi.org/10.1038/nature10450
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10450
This article is cited by
-
Identification of novel homozygous nonsense SLC10A7 variant causing short stature, amelogenesis imperfecta, and skeletal dysplasia with scoliosis and surgical management of spine
Orphanet Journal of Rare Diseases (2023)
-
Structural insight into the allosteric inhibition of human sodium-calcium exchanger NCX1 by XIP and SEA0400
The EMBO Journal (2023)
-
Structural insights into the HBV receptor and bile acid transporter NTCP
Nature (2022)
-
Structure of the bile acid transporter and HBV receptor NTCP
Nature (2022)
-
Structures and mechanism of the plant PIN-FORMED auxin transporter
Nature (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.