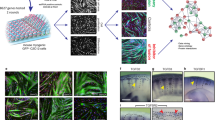
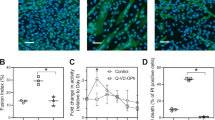
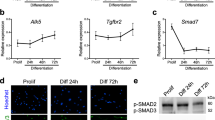
Abstract
Skeletal muscle arises from the fusion of precursor myoblasts into multinucleated myofibres1,2. Although conserved transcription factors and signalling proteins involved in myogenesis have been identified, upstream regulators are less well understood. Here we report an unexpected discovery that the membrane protein BAI1, previously linked to recognition of apoptotic cells by phagocytes3, promotes myoblast fusion. Endogenous BAI1 expression increased during myoblast fusion, and BAI1 overexpression enhanced myoblast fusion by means of signalling through ELMO/Dock180/Rac1 proteins4. During myoblast fusion, a fraction of myoblasts within the population underwent apoptosis and exposed phosphatidylserine, an established ligand for BAI1 (ref. 3). Blocking apoptosis potently impaired myoblast fusion, and adding back apoptotic myoblasts restored fusion. Furthermore, primary human myoblasts could be induced to form myotubes by adding apoptotic myoblasts, even under normal growth conditions. Mechanistically, apoptotic cells did not directly fuse with the healthy myoblasts, rather the apoptotic cells induced a contact-dependent signalling with neighbours to promote fusion among the healthy myoblasts. In vivo, myofibres from Bai1−/− mice are smaller than those from wild-type littermates. Muscle regeneration after injury was also impaired in Bai1−/− mice, highlighting a role for BAI1 in mammalian myogenesis. Collectively, these data identify apoptotic cells as a new type of cue that induces signalling via the phosphatidylserine receptor BAI1 to promote fusion of healthy myoblasts, with important implications for muscle development and repair.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Abmayr, S. M. & Pavlath, G. K. Myoblast fusion: lessons from flies and mice. Development 139, 641–656 (2012)
Chen, E. H. & Olson, E. N. Towards a molecular pathway for myoblast fusion in Drosophila . Trends Cell Biol. 14, 452–460 (2004)
Park, D. et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450, 430–434 (2007)
Gumienny, T. L. et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107, 27–41 (2001)
Hasegawa, H. et al. DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the cell membrane. Mol. Cell. Biol. 16, 1770–1776 (1996)
Brugnera, E. et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nature Cell Biol. 4, 574–582. (2002)
Erickson, M. R., Galletta, B. J. & Abmayr, S. M. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure, and cytoskeletal organization. J. Cell Biol. 138, 589–603 (1997)
Hakeda-Suzuki, S. et al. Rac function and regulation during Drosophila development. Nature 416, 438–442 (2002)
Moore, C. A., Parkin, C. A., Bidet, Y. & Ingham, P. W. A role for the Myoblast city homologues Dock1 and Dock5 and the adaptor proteins Crk and Crk-like in zebrafish myoblast fusion. Development 134, 3145–3153 (2007)
Laurin, M. et al. The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo . Proc. Natl Acad. Sci. USA 105, 15446–15451 (2008)
Vasyutina, E., Martarelli, B., Brakebusch, C., Wende, H. & Birchmeier, C. The small G-proteins Rac1 and Cdc42 are essential for myoblast fusion in the mouse. Proc. Natl Acad. Sci. USA 106, 8935–8940 (2009)
Geisbrecht, E. R. et al. Drosophila ELMO/CED-12 interacts with Myoblast city to direct myoblast fusion and ommatidial organization. Dev. Biol. 314, 137–149 (2008)
Yaffe, D. & Saxel, O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270, 725–727 (1977)
Cornelison, D. D. Context matters: in vivo and in vitro influences on muscle satellite cell activity. J. Cell. Biochem. 105, 663–669 (2008)
Pajcini, K. V., Pomerantz, J. H., Alkan, O., Doyonnas, R. & Blau, H. M. Myoblasts and macrophages share molecular components that contribute to cell-cell fusion. J. Cell Biol. 180, 1005–1019 (2008)
Shutes, A. et al. Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J. Biol. Chem. 282, 35666–35678 (2007)
van den Eijnde, S. M. et al. Transient expression of phosphatidylserine at cell-cell contact areas is required for myotube formation. J. Cell Sci. 114, 3631–3642 (2001)
Jeong, J. & Conboy, I. M. Phosphatidylserine directly and positively regulates fusion of myoblasts into myotubes. Biochem. Biophys. Res. Commun. 414, 9–13 (2011)
Chekeni, F. B. et al. Pannexin 1 channels mediate 'find-me' signal release and membrane permeability during apoptosis. Nature 467, 863–867 (2010)
Hasty, P. et al. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 364, 501–506 (1993)
Kamentsky, L. et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 27, 1179–1180 (2011)
Yan, Z. et al. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J. Biol. Chem. 278, 8826–8836 (2003)
Grounds, M. D., Radley, H. G., Lynch, G. S., Nagaraju, K. & De Luca, A. Towards developing standard operating procedures for pre-clinical testing in the mdx mouse model of Duchenne muscular dystrophy. Neurobiol. Dis. 31, 1–19 (2008)
Chen, Y. W., Zhao, P., Borup, R. & Hoffman, E. P. Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology. J. Cell Biol. 151, 1321–1336 (2000)
Haslett, J. N. et al. Gene expression comparison of biopsies from Duchenne muscular dystrophy (DMD) and normal skeletal muscle. Proc. Natl Acad. Sci. USA 99, 15000–15005 (2002)
Bakay, M. et al. Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb-MyoD pathways in muscle regeneration. Brain 129, 996–1013 (2006)
Bialek, P. et al. Distinct protein degradation profiles are induced by different disuse models of skeletal muscle atrophy. Physiol. Genomics 43, 1075–1086 (2011)
Fernando, P., Kelly, J. F., . Balazsi, K., Slack, R. S. & Megeney, L. A. Caspase 3 activity is required for skeletal muscle differentiation. Proc. Natl Acad. Sci. USA 99, 11025–11030 (2002)
Phaneuf, S. & Leeuwenburgh, C. Apoptosis and exercise. Med. Sci. Sports Exerc. 33, 393–396 (2001)
Ravichandran, K. S. Find-me and eat-me signals in apoptotic cell clearance: progress and conundrums. J. Exp. Med. 207, 1807–1817 (2010)
Sokolowski, J. D. et al. Brain-specific angiogenesis inhibitor-1 expression in astrocytes and neurons: implications for its dual function as an apoptotic engulfment receptor. Brain Behav. Immun. 25, 915–921 (2011)
Acknowledgements
We thank members of the Ravichandran laboratory for their valuable suggestions at many stages of this work. We also thank L. Haney, A. Bruce and A. Dutta for technical suggestions and assistance and M. Hufford and A. Fond for help with statistical analysis. We thank members of the University of Virginia Flow Cytometry Core, Research Histology Core, and Gene Targeting and Transgenic Facility for cell sorting, histological services and transgenic mouse generation. This work was supported by a grant from the National Institute of General Medical Sciences/National Institutes of Health and the Center for Cell Clearance at the University of Virginia.
Author information
Authors and Affiliations
Contributions
A.E.H.-H. designed, performed and analysed most of the experiments in this study with input from K.S.R. C.S.L. generated and supplied the Bai1−/− mice, and provided GST-tagged BAI1 TSR. J.M.K. assisted with time-lapse and shRNA studies, and myofibre cross-sectional area analyses. S.A. helped with the in vivo muscle regeneration and ex vivo primary myoblast cultures. J.D.S. processed, stained and analysed the mouse embryonic tissues. A.L.K. provided phosphatidylserine liposomes for these studies. J.A.C. and Z.Y. assisted with the cardiotoxin injury model. J.W.M. provided intellectual input on the in vivo BAI1 studies. A.E.H.-H. and K.S.R. wrote the manuscript with comments from co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-7. (PDF 39261 kb)
A population of myoblasts undergo cell death during fusion and remain in close contact with nascent myotubes
This video shows C2C12 fusing myoblasts labeled with TO-PRO-3 (red) to identify apoptotic cells. The time-lapse covers a period of 72 hours during which time a number of myoblasts die but do not appear to be phagocytosed. These TO-PRO-3 positive cells are passed along the newly forming myotubes culture maintain contact with the myotubes for the duration of the video. (MOV 28781 kb)
Rights and permissions
About this article
Cite this article
Hochreiter-Hufford, A., Lee, C., Kinchen, J. et al. Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature 497, 263–267 (2013). https://doi.org/10.1038/nature12135
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12135
This article is cited by
-
Anti-apoptotic protein Bcl-2 contributes to the determination of reserve cells during myogenic differentiation of C2C12 cells
In Vitro Cellular & Developmental Biology - Animal (2024)
-
Intrinsic signalling factors associated with cancer cell-cell fusion
Cell Communication and Signaling (2023)
-
Ghost messages: cell death signals spread
Cell Communication and Signaling (2023)
-
All-trans retinoic acid and dexamethasone regulate phagocytosis-related gene expression and enhance dead cell uptake in C2C12 myoblast cells
Scientific Reports (2023)
-
Biasing the conformation of ELMO2 reveals that myoblast fusion can be exploited to improve muscle regeneration
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.