Abstract
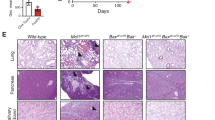
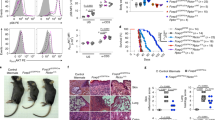
The mechanistic target of rapamycin (mTOR) pathway integrates diverse environmental inputs, including immune signals and metabolic cues, to direct T-cell fate decisions1. The activation of mTOR, which is the catalytic subunit of the mTORC1 and mTORC2 complexes, delivers an obligatory signal for the proper activation and differentiation of effector CD4+ T cells2,3, whereas in the regulatory T-cell (Treg) compartment, the Akt–mTOR axis is widely acknowledged as a crucial negative regulator of Treg-cell de novo differentiation4,5,6,7,8 and population expansion9. However, whether mTOR signalling affects the homeostasis and function of Treg cells remains largely unexplored. Here we show that mTORC1 signalling is a pivotal positive determinant of Treg-cell function in mice. Treg cells have elevated steady-state mTORC1 activity compared to naive T cells. Signals through the T-cell antigen receptor (TCR) and interleukin-2 (IL-2) provide major inputs for mTORC1 activation, which in turn programs the suppressive function of Treg cells. Disruption of mTORC1 through Treg-specific deletion of the essential component raptor leads to a profound loss of Treg-cell suppressive activity in vivo and the development of a fatal early onset inflammatory disorder. Mechanistically, raptor/mTORC1 signalling in Treg cells promotes cholesterol and lipid metabolism, with the mevalonate pathway particularly important for coordinating Treg-cell proliferation and upregulation of the suppressive molecules CTLA4 and ICOS to establish Treg-cell functional competency. By contrast, mTORC1 does not directly affect the expression of Foxp3 or anti- and pro-inflammatory cytokines in Treg cells, suggesting a non-conventional mechanism for Treg-cell functional regulation. Finally, we provide evidence that mTORC1 maintains Treg-cell function partly through inhibiting the mTORC2 pathway. Our results demonstrate that mTORC1 acts as a fundamental ‘rheostat’ in Treg cells to link immunological signals from TCR and IL-2 to lipogenic pathways and functional fitness, and highlight a central role of metabolic programming of Treg-cell suppressive activity in immune homeostasis and tolerance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Chi, H. Regulation and function of mTOR signalling in T cell fate decisions. Nature Rev. Immunol. 12, 325–338 (2012)
Delgoffe, G. M. et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nature Immunol. 12, 295–303 (2011)
Lee, K. et al. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity 32, 743–753 (2010)
Delgoffe, G. M. et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity 30, 832–844 (2009)
Haxhinasto, S., Mathis, D. & Benoist, C. The AKT–mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J. Exp. Med. 205, 565–574 (2008)
Sauer, S. et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc. Natl Acad. Sci. USA 105, 7797–7802 (2008)
Liu, G. et al. The receptor S1P1 overrides regulatory T cell-mediated immune suppression through Akt-mTOR. Nature Immunol. 10, 769–777 (2009)
Liu, G., Yang, K., Burns, S., Shrestha, S. & Chi, H. The S1P1-mTOR axis directs the reciprocal differentiation of TH1 and Treg cells. Nature Immunol. 11, 1047–1056 (2010)
Battaglia, M., Stabilini, A. & Roncarolo, M. G. Rapamycin selectively expands CD4+CD25+FoxP3+ regulatory T cells. Blood 105, 4743–4748 (2005)
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012)
Procaccini, C. et al. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity 33, 929–941 (2010)
Kelly, A. P. et al. Notch-induced T cell development requires phosphoinositide-dependent kinase 1. EMBO J. 26, 3441–3450 (2007)
Cunningham, J. T. et al. mTOR controls mitochondrial oxidative function through a YY1–PGC-1α transcriptional complex. Nature 450, 736–740 (2007)
van der Windt, G. J. et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012)
Gurumurthy, S. et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature 468, 659–663 (2010)
Takahashi, T. et al. Immunologic self-tolerance maintained by CD25+CD4+ naturally anergic and suppressive T cells: induction of autoimmune disease by breaking their anergic/suppressive state. Int. Immunol. 10, 1969–1980 (1998)
Fisson, S. et al. Continuous activation of autoreactive CD4+ CD25+ regulatory T cells in the steady state. J. Exp. Med. 198, 737–746 (2003)
Gavin, M. A., Clarke, S. R., Negrou, E., Gallegos, A. & Rudensky, A. Homeostasis and anergy of CD4+CD25+ suppressor T cells in vivo. Nature Immunol. 3, 33–41 (2002)
Thornton, A. M., Donovan, E. E., Piccirillo, C. A. & Shevach, E. M. Cutting edge: IL-2 is critically required for the in vitro activation of CD4+CD25+ T cell suppressor function. J. Immunol. 172, 6519–6523 (2004)
Herman, A. E., Freeman, G. J., Mathis, D. & Benoist, C. CD4+CD25+ T regulatory cells dependent on ICOS promote regulation of effector cells in the prediabetic lesion. J. Exp. Med. 199, 1479–1489 (2004)
Wing, K. et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science 322, 271–275 (2008)
Rubtsov, Y. P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008)
Kanangat, S. et al. Disease in the scurfy (sf) mouse is associated with overexpression of cytokine genes. Eur. J. Immunol. 26, 161–165 (1996)
Chaudhry, A. et al. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity 34, 566–578 (2011)
Josefowicz, S. Z., Lu, L. F. & Rudensky, A. Y. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564 (2012)
Ouyang, W. et al. Novel Foxo1-dependent transcriptional programs control Treg cell function. Nature 491, 554–559 (2012)
Wang, R. & Green, D. R. Metabolic checkpoints in activated T cells. Nature Immunol. 13, 907–915 (2012)
Tai, X. et al. Basis of CTLA-4 function in regulatory and conventional CD4+ T cells. Blood 119, 5155–5163 (2012)
Corse, E. & Allison, J. P. Cutting edge: CTLA-4 on effector T cells inhibits in trans. J. Immunol. 189, 1123–1127 (2012)
Sadlack, B. et al. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell 75, 253–261 (1993)
Yang, K., Neale, G., Green, D. R., He, W. & Chi, H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nature Immunol. 12, 888–897 (2011)
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005)
Acknowledgements
We thank A. Rudensky for Foxp3YFP-Cre mice, D. Green for help with metabolic assays, N. Brydon for animal colony management, Y. Wang for editing of the manuscript, and the St Jude Immunology FACS core facility for cell sorting. This work was supported by the US National Institutes of Health (K01 AR053573, R21 AI094089, R01 AI101407 and R01 NS064599), the Lupus Research Institute, and the American Lebanese Syrian Associated Charities (all to H.C.).
Author information
Authors and Affiliations
Contributions
H.Z. designed and performed experiments, and wrote the manuscript; K.Y. contributed to cellular experiments; C.C. contributed to survival curves and technical support; G.N. performed bioinformatic analyses; P.V. performed histological analysis; H.C. designed experiments, contributed to writing the manuscript, and provided overall direction.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-17. (PDF 1314 kb)
Rights and permissions
About this article
Cite this article
Zeng, H., Yang, K., Cloer, C. et al. mTORC1 couples immune signals and metabolic programming to establish Treg-cell function. Nature 499, 485–490 (2013). https://doi.org/10.1038/nature12297
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12297
This article is cited by
-
Immunosurveillance encounters cancer metabolism
EMBO Reports (2024)
-
The regulation and differentiation of regulatory T cells and their dysfunction in autoimmune diseases
Nature Reviews Immunology (2024)
-
The mechanism of dendritic cell-T cell crosstalk in rheumatoid arthritis
Arthritis Research & Therapy (2023)
-
Cholesterol suppresses human iTreg differentiation and nTreg function through mitochondria-related mechanisms
Journal of Translational Medicine (2023)
-
Lipid metabolic reprogramming in tumor microenvironment: from mechanisms to therapeutics
Journal of Hematology & Oncology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.