Abstract
Interactions between species can promote evolutionary divergence of ecological traits and social signals1,2, a process widely assumed to generate species differences in adaptive radiation3,4,5. However, an alternative view is that lineages typically interact when relatively old6, by which time selection for divergence is weak7,8 and potentially exceeded by convergent selection acting on traits mediating interspecific competition9. Few studies have tested these contrasting predictions across large radiations, or by controlling for evolutionary time. Thus the role of species interactions in driving broad-scale patterns of trait divergence is unclear10. Here we use phylogenetic estimates of divergence times to show that increased trait differences among coexisting lineages of ovenbirds (Furnariidae) are explained by their greater evolutionary age in relation to non-interacting lineages, and that—when these temporal biases are accounted for—the only significant effect of coexistence is convergence in a social signal (song). Our results conflict with the conventional view that coexistence promotes trait divergence among co-occurring organisms at macroevolutionary scales, and instead provide evidence that species interactions can drive phenotypic convergence across entire radiations, a pattern generally concealed by biases in age.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Data deposits
Nuclear and mitochondrial DNA sequences for all lineages have been deposited in GenBank under accession numbers given in Supplementary Data 1.
References
Grant, P. R. & Grant, B. R. Evolution of character displacement in Darwin’s finches. Science 313, 224–226 (2006)
Grant, B. R. & Grant, P. R. Songs of Darwin’s finches diverge when a new species enters the community. Proc. Natl Acad. Sci. USA 107, 20156–20163 (2010)
Darwin, C. R. On the Origin of Species (John Murray, 1859)
Simpson, G. G. Tempo and Mode in Evolution (Columbia Univ. Press, 1944)
Pfennig, D. W. & Pfennig, K. S. Character displacement and the origins of diversity. Am. Nat. 176, S26–S44 (2010)
Weir, J. T. & Price, T. D. Limits to speciation inferred from times to secondary sympatry and ages of hybridizing species along a latitudinal gradient. Am. Nat. 177, 462–469 (2011)
Price, T. D. The roles of time and ecology in the continental radiation of the Old World leaf warblers (Phylloscopus and Seicercus). Phil. Trans. R. Soc. B 365, 1749–1762 (2010)
Seddon, N. & Tobias, J. A. Character displacement from the receiver’s perspective: species and mate recognition despite convergent signals in suboscine birds. Proc. R. Soc. Lond. B 277, 2475–2483 (2010)
Grether, G. F., Losin, N., Anderson, C. N. & Okamoto, K. The role of interspecific interference competition in character displacement and the evolution of competitor recognition. Biol. Rev. Camb. Philos. Soc. 84, 617–635 (2009)
Reznick, D. N. & Ricklefs, R. E. Darwin’s bridge between microevolution and macroevolution. Nature 457, 837–842 (2009)
Brown, W. L. & Wilson, E. O. Character displacement. Syst. Zool. 5, 49–64 (1956)
Pfennig, K. S. & Pfennig, D. W. Character displacement: ecological and reproductive responses to a common evolutionary problem. Q. Rev. Biol. 84, 253–276 (2009)
Gavrilets, S. & Losos, J. B. Adaptive radiation: contrasting theory with data. Science 323, 732–737 (2009)
Dayan, T. & Simberloff, D. Ecological and community-wide character displacement: the next generation. Ecol. Lett. 8, 875–894 (2005)
Schoener, T. W. The evolution of bill size differences among sympatric congeneric species of birds. Evolution 19, 189–213 (1965)
Sætre, G. P. et al. A sexually selected character displacement in flycatchers reinforces premating isolation. Nature 387, 589–592 (1997)
Davies, T., Meiri, S., Barraclough, T. & Gittleman, J. Species co-existence and character divergence across carnivores. Ecol. Lett. 10, 146–152 (2007)
Connell, J. H. Diversity and the coevolution of competitors, or the ghost of competition past. Oikos 35, 131–138 (1980)
Rundell, R. J. & Price, T. D. Adaptive radiation, non-adaptive radiation, ecological speciation and non-ecological speciation. Trends Ecol. Evol. 24, 394–399 (2009)
Derryberry, E. P. et al. Lineage diversification and morphological evolution in a large-scale continental radiation: the Neotropical ovenbirds and woodcreepers (Aves: Furnariidae). Evolution 65, 2973–2986 (2011)
Seddon, N. Ecological adaptation and species recognition drive vocal evolution in Neotropical suboscine birds. Evolution 59, 200–215 (2005)
Tobias, J. A. et al. Song divergence by sensory drive in Amazonian birds. Evolution 64, 2820–2839 (2010)
Derryberry, E. P. et al. Correlated evolution of beak morphology and song in the Neotropical woodcreeper radiation. Evolution 66, 2784–2797 (2012)
Huntley, J. W. et al. Testing limiting similarity in Quaternary terrestrial gastropods. Paleobiology 34, 378–388 (2008)
Monroe, M. J. Does competition drive character differences between species on a macroevolutionary scale? J. Evol. Biol. 25, 2341–2347 (2012)
Pigot, A. L. & Tobias, J. A. Species interactions constrain geographic range expansion over evolutionary time. Ecol. Lett. 16, 330–338 (2012)
Tobias, J. A. & Seddon, N. Signal design and perception in Hypocnemis antbirds: evidence for convergent evolution via social selection. Evolution 63, 3168–3189 (2009)
Laiolo, P. Interspecific interactions drive cultural co-evolution and acoustic convergence in syntopic species. J. Anim. Ecol. 81, 594–604 (2012)
Weir, J. T., Wheatcroft, D. & Price, T. The role of ecological constraint in driving the evolution of avian song frequency across a latitudinal gradient. Evolution 66, 2773–2783 (2012)
Muschick, M., Indermaur, A. & Salzburger, W. Convergent evolution within an adaptive radiation of cichlid fishes. Curr. Biol. 22, 2362–2368 (2012)
Ridgely, R. S. et al. Digital Distribution Maps of the Birds of the Western Hemisphere, v. 2.1 (NatureServe, 2005)
Chesser, R. T. & Zink, R. M. Mode of speciation in birds: a test of Lynch’s method. Evolution 48, 490–497 (1995)
Lynch, J. D. in Speciation and its Consequences (eds Otte, D. & Endler, J. A. ) 527–553 (Sinauer, 1989)
Kenward, M. G. & Roger, J. H. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics 53, 983–997 (1997)
Self, S. G. & Liang, K.-Y. Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J. Am. Stat. Assoc. 82, 605–610 (1987)
Gilmour, A. R., Gogel, B. J., Cullis, B. R., Welham, S. J. & Thompson, R. ASReml User Guide, Release 3.0 (VSN International, 2009)
Hadfield, J. D. & Nakagawa, S. General quantitative genetic methods for comparative biology: phylogenies, taxonomies, meta-analysis and multi-trait models for continuous and categorical characters. J. Evol. Biol. 23, 494–508 (2010)
Paradis, E., Claude, J. & Strimmer, K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004)
Pinheiro, J., Bates, D., DebRoy, S. & Sarkar, D. Package nlme: Linear and Nonlinear Mixed Effects Models, v. 3.1-109 (R-core, 2013)
Harmon, L. J., Weir, J. T., Brock, C. D., Glor, R. E. & Challenger, W. GEIGER: investigating evolutionary radiations. Bioinformatics 24, 129–131 (2008)
Acknowledgements
We thank G. Grether, J. Hadfield, S. Nakagawa, A. Phillimore, A. Pigot, R. Ricklefs, G. Thomas and S. West for comments and discussion. We are also indebted to the many individuals who collected specimens, tissue samples and sound recordings, and to numerous institutions (particularly the Macaulay Library, Cornell University) for granting access to this material. Complete acknowledgements and data sets are provided in the Supplementary Information. This research was supported by the John Fell Fund (to J.A.T.), the Browne Fellowship, Queen’s College, Oxford, and Vetenskapsrådet (to C.K.C.), the National Science Foundation (to R.T.B.) and the Royal Society (to N.S.).
Author information
Authors and Affiliations
Contributions
J.A.T. and N.S. conceived and designed the study, compiled and analysed song data, and integrated all data sets; S.C. provided morphometric data; E.P.D., S.C. and R.T.B. conducted molecular sequencing and phylogenetic analyses; C.C. designed and conducted statistical analyses, with significant input from N.S.; N.S., J.A.T. and C.C. produced figures and tables; J.A.T. prepared and edited the manuscript, with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
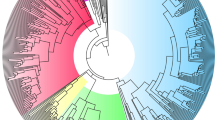
Extended Data Figure 1 Phenotypic radiation.
Variation in beak morphology and plumage across 350 lineages in 12 major clades of ovenbirds (species richness in each clade is represented by distance on the circumference of the phylogram; clades are coloured to facilitate interpretation). Numbered clades (species names running clockwise) are as follows: 1, Synallaxini, clade A (Certhiaxis cinnamomeus, Pseudoseisura cristata, Synallaxis azarae, Pseudasthenes cactorum); 2, Synallaxini, clade B (Cranioleuca hellmayri, Acrobatornis fonsecai, Thripophaga macroura); 3, Synallaxini, clade C (Asthenes pyrrholeuca, Coryphistera alaudina); 4, Phacellodomus (Phacellodomus erythrophthalmus); 5, Leptasthenura (Leptasthenura xenothorax, Aphrastura spinicauda); 6, Margarornis/Premnoplex (Margarornis squamiger); 7, Philydorini (Automolus ochrolaemus, Thripadectes flammulatus, Syndactyla ucayalae, Hylocryptus erythrocephalis, Clibanornis dendrocolaptoides); 8, Furnariini (Furnarius cristatus, Upucerthia jelskii); 9, Ochetorhynchus (Pygarrhichas albogularis); 10, Xenops (Xenops tenuirostris); 11, Dendrocolaptinae (Nasica longirostris, Sittasomus griseicapillus, Campylorhamphus trochilirostris, Hylexetastes stresemanni, Dendroplex picus); 12, Sclerurinae (Geositta tenuirostris, Geositta antarctica, Sclerurus guatemalensis). Illustrations reproduced with the permission of Lynx Edicions.
Extended Data Figure 2 Stereotypy in ovenbird songs.
Examples of single songs given by six different individuals of one species of ovenbird (Xiphorhynchus guttatus) to illustrate typical levels of stereotypy of acoustic structure in a song sample in our data set, as well as the relative simplicity of suboscine songs (compared with the songs of oscine songbirds). Songs recorded in Bolivia (a–c) and Peru (d–f) were used to generate spectrograms (time versus frequency) using Raven Pro (settings: window = Hann, bandwidth = 256 Hz, fast Fourier transform = 1,024, overlap = 0.875). Catalogue numbers of recordings: (a) XC2297, (b) XC64610, (c) XC1756, (d) JATXiphGutt02, (e) JATXiphGutt08, (f) XC84 (see Supplementary Data 1).
Extended Data Figure 3 Representative songs from the seven major clades of ovenbirds.
a, Synallaxini, clade A: Synallaxis erythrothorax (catalogue number ML7048, country El Salvador); b, Synallaxini, clade B: Cranioleuca curtata (ML120989, Bolivia); c, Synallaxini, clade C: Asthenes modesta (JAT99129, Bolivia); d, Philydorini: Automolus rufipileatus consobrina (ML66235, Venezuela); e, Furnariini: Cinclodes aricomae (JAT99138, Bolivia); f, Dendrocolaptinae: Campylorhamphus trochilirostris hellmayri (XC48901, Argentina); g, Sclerurinae: Geositta rufipennis rufipennis (ML46401, Argentina). Shown are waveforms (time versus amplitude, above) and spectrograms (time versus frequency) generated in MATLAB. Vertical dashed red lines (visible at high magnification) show note onset and offset.
Extended Data Figure 4 Evolutionary ages of ovenbird lineages.
Histograms show the frequency distribution of ages in allopatry (blue: a, b) versus sympatry (green: c, d) for pairs of closest relatives (a, c) and for all pairs of lineages (b, d). The reduced age of allopatric relatives is most evident in a: only seven (2.6%) of the closest sympatric lineages were estimated to be <2 Myr old as opposed to 79 (31.6%) of closest allopatric lineages. Data are from all 350 lineages, excluding pairwise comparisons across subfamilies (that is, restricting to comparisons in Furnariinae, Dendrocolaptinae and Sclerurinae).
Extended Data Figure 5 Relationship between habitat divergence and evolutionary age in ovenbirds.
Histograms show patterns in allopatry (blue: a–c) versus sympatry (green: d–f), plotted against evolutionary age: a, d, lineages occupying the same habitat (that is, both in closed, semi-open or open habitats); b, e, lineages occupying moderately divergent habitats (closed versus semi-open, or semi-open versus open); c, f, lineages occupying very different habitats (closed versus open). Data are from all 350 lineages, excluding pairwise comparisons across subfamilies (that is, restricting to comparisons in Furnariinae, Dendrocolaptinae and Sclerurinae).
Extended Data Figure 6 Permutation tests examining the influence of data structure and response variable distribution on fixed effects.
Histograms show the frequency distributions of parameter estimates (x axis) obtained by randomly shuffling the response variable and re-running models 1,000 times. This procedure was applied to each fixed effect included in PLMMs, first on all pairs of closest relatives (upper panels) and then on all lineages (lower panels). Red lines show the parameter estimates obtained in our main analyses on observed data; P values refer to the proportion of permutations for which the parameter estimate obtained from observed data was greater (or less) than the randomly generated response. In all analyses, permutation P values were very similar to those obtained using F-tests (close relatives: Supplementary Tables 4–6; all lineages: Supplementary Tables 18–20), confirming that our results were not explained by biases in the structure of our data sets and variation in the distribution of our response variables.
Extended Data Figure 7 Comparing observed and simulated data sets under best-fit models of evolution.
Histograms show estimated differences between allopatric and sympatric lineages of ovenbird in beak morphology (a, b), tarsus length (c, d) and song structure (e, f), comparing values obtained in our main analyses (red line; Supplementary Tables 4–6 and 18–20) with data simulated under best-fit models of evolution. Different colours denote simulated trait data following a Brownian motion model of evolution (blue), and an Ornstein–Uhlenbeck model (green). P values are the proportion of simulations (n = 1,000) where the estimated difference obtained from our main analysis was greater (or less) than the estimate from the simulated data. For all traits and data sets, we found that P values from simulations corresponded to those in main analyses. Note that convergence in beaks is marginally significant and restricted to the all-lineages analysis (b), whereas convergence in songs is strongly significant and consistent across both analyses (e, f).
Extended Data Figure 8 Recovery of simulated trait divergence under different models of trait evolution.
Plots compare simulated and observed trait divergence between pairs of closely related ovenbird lineages (allopatric and sympatric lineages pooled) under two models of evolution using PLMMs: a, Brownian motion; b, Ornstein–Uhlenbeck. These 1:1 relationships demonstrate that PLMMs were able to recover differences simulated under both evolutionary models detected in our data sets. Scatter is reduced under Ornstein–Uhlenbeck because divergence is constrained.
Extended Data Figure 9 Examples of ovenbird lineages with similar songs in sympatry.
a, Synallaxis frontalis and b, Synallaxis azarae superciliosus; c, Dendrocolaptes picumnus picumnus and d, Dendrocolaptes certhia concolor; e, Dendroplex picus and f, Dendroplex kieneri. For each pair of recordings (taken at the same locality), spectrograms show that songs of each lineage are species specific, but highly similar in key acoustic traits (for example, peak frequency or pace) to their closest sympatric relative. Figures are annotated with the approximate time in millions of years since divergence. Illustrations reproduced with the permission of Lynx Edicions.
Extended Data Figure 10 Interpreting linear and quadratic terms of evolutionary age.
Trait data were simulated over 100 randomly generated trees specifying four different models of evolution: a, Brownian motion; b, Ornstein–Uhlenbeck (α = 0.1); c, Ornstein–Uhlenbeck (α = 0.3); d, Ornstein–Uhlenbeck (α = 0.6). Pairwise genetic (x axis: evolutionary age) and trait differences (y axis: trait divergence) between taxa were extracted from simulated data. Trait data and trees were simulated using the R package ‘ape’ with the functions rTraitCont and rtree. Plots show that the Brownian motion model predicts a linear term and the Ornstein–Uhlenbeck model predicts a quadratic term, particularly when the constraint parameter increases in strength.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Tables 1-27, a Supplementary Discussion, Supplementary Notes, Supplementary References and a Supplementary Code for Statistical Analyses. (PDF 2378 kb)
Supplementary Data
The file contains Supplementary Datasets 1-7 – see file for details. (XLSX 259 kb)
Rights and permissions
About this article
Cite this article
Tobias, J., Cornwallis, C., Derryberry, E. et al. Species coexistence and the dynamics of phenotypic evolution in adaptive radiation. Nature 506, 359–363 (2014). https://doi.org/10.1038/nature12874
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12874
This article is cited by
-
Mosaic Evolution of Grasping and Genital Traits in Two Sympatric Scorpion Species with Reproductive Interference
Evolutionary Biology (2024)
-
Conceptual and empirical bridges between micro- and macroevolution
Nature Ecology & Evolution (2023)
-
Number of forest fragments and understory plants exert opposite effects on multiple facets of bird diversity in eucalypt plantations
Landscape Ecology (2023)
-
Environmental signal in the evolutionary diversification of bird skeletons
Nature (2022)
-
Learning in advance? Interspecific recognition ability in male Eurasian blackcaps
Journal of Ornithology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.