Abstract
Infectious agents develop intricate mechanisms to interact with host cell pathways and hijack their genetic and epigenetic machinery to change host cell phenotypic states. Among the Apicomplexa phylum of obligate intracellular parasites, which cause veterinary and human diseases, Theileria is the only genus that transforms its mammalian host cells1. Theileria infection of bovine leukocytes induces proliferative and invasive phenotypes associated with activated signalling pathways, notably JNK and AP-1 (ref. 2). The transformed phenotypes are reversed by treatment with the theilericidal drug buparvaquone3. We used comparative genomics to identify a homologue of the peptidyl-prolyl isomerase PIN1 in T. annulata (TaPIN1) that is secreted into the host cell and modulates oncogenic signalling pathways. Here we show that TaPIN1 is a bona fide prolyl isomerase and that it interacts with the host ubiquitin ligase FBW7, leading to its degradation and subsequent stabilization of c-JUN, which promotes transformation. We performed in vitro and in silico analysis and in vivo zebrafish xenograft experiments to demonstrate that TaPIN1 is directly inhibited by the anti-parasite drug buparvaquone (and other known PIN1 inhibitors) and is mutated in a drug-resistant strain. Prolyl isomerization is thus a conserved mechanism that is important in cancer and is used by Theileria parasites to manipulate host oncogenic signalling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dobbelaere, D. & Heussler, V. Transformation of leukocytes by Theileria parva and T. annulata . Annu. Rev. Microbiol. 53, 1–42 (1999)
Chaussepied, M. et al. Upregulation of Jun and Fos family members and permanent JNK activity lead to constitutive AP-1 activation in Theileria-transformed leukocytes. Mol. Biochem. Parasitol. 94, 215–226 (1998)
McHardy, N., Wekesa, L. S., Hudson, A. T. & Randall, A. W. Antitheilerial activity of BW720C (buparvaquone): a comparison with parvaquone. Res. Vet. Sci. 39, 29–33 (1985)
Shiels, B. R. et al. A Theileria annulata DNA binding protein localized to the host cell nucleus alters the phenotype of a bovine macrophage cell line. Eukaryot. Cell 3, 495–505 (2004)
Pain, A. et al. Genome of the host-cell transforming parasite Theileria annulata compared with T. parva . Science 309, 131–133 (2005)
Witschi, M. et al. Proteomic analysis of the Theileria annulata schizont. Int. J. Parasitol. 43, 173–180 (2013)
Lu, K. P., Hanes, S. D. & Hunter, T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 380, 544–547 (1996)
Winkler, K. E., Swenson, K. I., Kornbluth, S. & Means, A. R. Requirement of the prolyl isomerase Pin1 for the replication checkpoint. Science 287, 1644–1647 (2000)
Wulf, G. M. et al. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 20, 3459–3472 (2001)
Ryo, A. et al. PIN1 is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells. Mol. Cell. Biol. 22, 5281–5295 (2002)
Yaffe, M. B. et al. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 278, 1957–1960 (1997)
Lu, K. P., Finn, G., Lee, T. H. & Nicholson, L. K. Prolyl cis-trans isomerization as a molecular timer. Nature Chem. Biol. 3, 619–629 (2007)
Hennig, L. et al. Selective Inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry 37, 5953–5960 (1998)
Tatara, Y., Lin, Y.-C., Bamba, Y., Mori, T. & Uchida, T. Dipentamethylene thiuram monosulfide is a novel inhibitor of Pin1. Biochem. Biophys. Res. Commun. 384, 394–398 (2009)
Moore, J. D. & Potter, A. Pin1 inhibitors: pitfalls, progress and cellular pharmacology. Bioorg. Med. Chem. Lett. 23, 4283–4291 (2013)
Hayashida, K. et al. Comparative genome analysis of three eukaryotic parasites with differing abilities to transform leukocytes reveals key mediators of Theileria-induced leukocyte transformation. MBio 3, e00204–12 (2012)
Chen, C.-H. et al. SENP1 desumoylates and regulates Pin1 protein activity and cellular function. Cancer Res. 73, 3951–3962 (2013)
Landrieu, I. et al. The Arabidopsis thaliana PIN1At gene encodes a single-domain phosphorylation-dependent peptidyl prolyl cis/trans isomerase. J. Biol. Chem. 275, 10577–10581 (2000)
Landrieu, I., Wieruszeski, J.-M., Wintjens, R., Inzé, D. & Lippens, G. Solution structure of the single-domain prolyl cis/trans isomerase PIN1At from Arabidopsis thaliana . J. Mol. Biol. 320, 321–332 (2002)
Yao, J.-L., Kops, O., Lu, P.-J. & Lu, K. P. Functional conservation of phosphorylation-specific prolyl isomerases in plants. J. Biol. Chem. 276, 13517–13523 (2001)
Goh, J. Y. et al. Functional characterization of two novel parvulins in Trypanosoma brucei . FEBS Lett. 584, 2901–2908 (2010)
Sun, L. et al. Solution structural analysis of the single-domain parvulin TbPin1. PLoS ONE 7, e43017 (2012)
Mhadhbi, M. et al. In vivo evidence for the resistance of Theileria annulata to buparvaquone. Vet. Parasitol. 169, 241–247 (2010)
Sharifiyazdi, H., Namazi, F., Oryan, A., Shahriari, R. & Razavi, M. Point mutations in the Theileria annulata cytochrome b gene is associated with buparvaquone treatment failure. Vet. Parasitol. 187, 431–435 (2012)
Konantz, M. et al. Zebrafish xenografts as a tool for in vivo studies on human cancer. Ann. NY Acad. Sci. 1266, 124–137 (2012)
White, R., Rose, K. & Zon, L. Zebrafish cancer: the state of the art and the path forward. Nature Rev. Cancer 13, 624–636 (2013)
Min, S.-H. et al. Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase. Mol. Cell 46, 771–783 (2012)
Nateri, A. S., Riera-Sans, L., Costa, C. D. & Behrens, A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 303, 1374–1378 (2004)
Wei, W., Jin, J., Schlisio, S., Harper, J. W. & Kaelin, W. G. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8, 25–33 (2005)
Marsolier, J. et al. OncomiR addiction is generated by a miR-155 feedback loop in Theileria-transformed leukocytes. PLoS Pathog. 9, e1003222 (2013)
Moreau, M.-F. et al. Theileria annulata in CD5+ macrophages and B1 B cells. Infect. Immun. 67, 6678–6682 (1999)
Aurrecoechea, C. et al. ApiDB: integrated resources for the apicomplexan bioinformatics resource center. Nucleic Acids Res. 35, D427–D430 (2007)
Lambert, C., Léonard, N., Bolle, X. D. & Depiereux, E. ESyPred3D: prediction of proteins 3D structures. Bioinformatics 18, 1250–1256 (2002)
Ranganathan, R., Lu, K. P., Hunter, T. & Noel, J. P. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell 89, 875–886 (1997)
Miteva, M. A., Tufféry, P. & Villoutreix, B. O. PCE: web tools to compute protein continuum electrostatics. Nucleic Acids Res. 33, W372–W375 (2005)
Grove, L. E., Hall, D. R., Beglov, D., Vajda, S. & Kozakov, D. FTFlex: accounting for binding site flexibility to improve fragment-based identification of druggable hot spots. Bioinformatics 29, 1218–1219 (2013)
Lagorce, D., Pencheva, T., Villoutreix, B. O. & Miteva, M. A. DG-AMMOS: a new tool to generate 3D conformation of small molecules using distance geometry and automated molecular mechanics optimization for in silico screening. BMC Chem. Biol. 9, 6 (2009)
Spitzer, R. & Jain, A. N. Surflex-Dock: docking benchmarks and real-world application. J. Comput. Aided Mol. Des. 26, 687–699 (2012)
Thomsen, R. & Christensen, M. H. MolDock: a new technique for high-accuracy molecular docking. J. Med. Chem. 49, 3315–3321 (2006)
Sauton, N., Lagorce, D., Villoutreix, B. O. & Miteva, M. A. MS-DOCK: accurate multiple conformation generator and rigid docking protocol for multi-step virtual ligand screening. BMC Bioinformatics 9, 184 (2008)
Ji, Q. et al. Molecular mechanism of quinone signaling mediated through S-quinonization of a YodB family repressor QsrR. Proc. Natl Acad. Sci. USA 110, 5010–5015 (2013)
Acknowledgements
We thank G. Langsley, M. A. Darghouth and M. Weitzman for critical reading of the manuscript and advice on this study. We thank members of the UMR7216 for discussions. J.B.W. thanks C. Gawer for advice and support. We thank the following for providing reagents: G. Langsley for TBL3-, BL3- and TpMD409-infected cells and for a Theileria complementary DNA library; C. Francastel for NIH/3T3 cells; G. Del Sal for Pin1−/− murine immortalized fibroblasts; B. E. Clurman for FBW7 plasmids; J. Baum for the rabbit anti-Apicomplexa actin antibody; and T. Uchida for DTM. Measurements of the PPIase activities were performed at the Flexstation III facility of the Biologie Fonctionnelle et Adaptative laboratory. Confocal analysis was performed at the microscopy facility of the ImagoSeine platform (Jacques Monod Institute). This work was supported by National Institutes of Health grant R01CA167677 to K.P.L., the Association for International Cancer Research (#08-0111), the French National Research Agency (ANR) (Blanc 11-BSV3-016-01), and the “Who Am I?” Laboratory of Excellence #ANR-11-LABX-0071 funded by the French Government through its “Investments for the Future” program operated by the ANR under grant #ANR-11-IDEX-0005-01.
Author information
Authors and Affiliations
Contributions
J.M. and M.P. performed the experiments; J.D.D. performed the comparative genomics bioinformatics screen; B.O.V. performed the structural three-dimensional modelling; J.C., T.L. and C.G. designed and executed the zebrafish experiments; L.F., S.A.-S.-A., X.Z.Z. and K.P.L. provided critical reagents and advice; M.M. provided genomic DNA from buparvaquone-resistant parasites; J.M., S.M. and J.B.W. conceived the study and designed experiments, analysed the data and wrote the paper. All authors read the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Bioinformatic screen and in silico predicted signal peptide for TaPIN1.
a, On 10 September 2011, search strategies at EuPathDB were used to search for all T. annulata protein-encoding genes with predicted signal peptides (SignalP 2.0 HMM). Six-hundred and eighty-nine genes were returned. One-hundred and thirty-eight of these were found to have a predicted signal peptide only in T. annulata and not in their T. gondii orthologues. Among these proteins, we excluded (1) hypothetical proteins, (2) proteins that are not expressed at the macroschizont stage, as the macroschizont is the stage of the parasite associated with host cell transformation, and (3) proteins that are predicted to be targeted to the apicoplast of the parasite. We obtained 33 proteins (right panel). b, Sequence alignment of PIN1 proteins in H. sapiens (human), B. taurus (cow), M. musculus (mouse) and T. annulata revealed a predicted signal peptide (underlined) and conserved PPIase domain in TaPIN1. Stars indicate TaPIN1 residues mutated in the study. c, Example of signal peptide predicted with SignaIP 3.0 Server.
Extended Data Figure 2 TaPIN1 is conserved between T. annulata and T. parva.
a, As the T. parva TpPIN1 sequence did not have an annotated signal peptide, we reanalysed sequence alignment of pin1 genes in T. annulata (Tapin1) and T. parva (Tppin1) (National Center for Biotechnology Information (NCBI) BLAST). The annotated T. annulata intron is indicated in grey and the annotated methionine codon (ATG) in T. parva is in black (underlined). Tapin1 and Tppin1 are well conserved, including in the intronic sequence. We conclude that the T. parva Tppin1 annotation should be modified in light of our polymerase chain reaction (PCR) and western blot analysis (see Fig. 1b and Extended Data Fig. 5c). b, Sequence alignment of pin1 genes in T. annulata (Tapin1) and T. orientalis (Topin1) (NCBI BLAST). The annotated T. annulata intron is indicated in grey and the annotated methionine codon in T. orientalis is in black (underlined). Tapin1 and Topin1 are less conserved than Tapin1 and Tppin1.
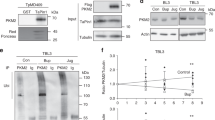
Extended Data Figure 3 BtPin1 is not regulated by the parasite.
a, b, Buparvaquone (Bup) treatment had no effect on the mRNA levels (a) of bovine Pin1 in infected (TBL3) or non-infected (BL3) cells as assessed by qPCR analysis, or on the protein level (b) as assessed by immunoblot analysis. Con, control. Average ± s.d., n = 3. b, The result is representative of three independent experiments.
Extended Data Figure 4 Characterization of the TaPIN1 antibody and secretion of TpPIN1 in T. parva.
a, The antibody raised against TaPIN1 specifically recognizes the Theileria protein in NIH/3T3 cells stably transfected with TaPIN1 but not the empty vector in NIH/3T3 cells stably transfected with control or the human protein in NIH/3T3 cells stably transfected with hPIN1. Both Theileria and human versions of PIN1 are present in NIH/3T3 cells stably transfected with TaPIN1 and NIH/3T3 cells stably transfected with hPIN1, respectively. b, The antibody raised against TaPIN1 specifically recognizes GST–TaPIN1 but not GST and GST–hPIN1 beads. c, TpPIN1 protein was detected in the nuclear and cytoplasmic fractions of T. parva-infected TpMD409 cells and decreased upon buparvaquone (Bup) treatment. Con, control. Antibodies recognizing apicomplexan actin, bovine tubulin or bovine histone H3 were used as controls. d, TaPIN1 expression in BL3 or parasite-infected TBL3 cells was examined by confocal microscopy analysis using an antibody raised specifically against Theileria TaPIN1, counterstaining with DAPI. Three-dimensional reconstructions in BL3 and TBL3 cells are shown. e, Immunostaining of TaPIN1 and haemagglutin tag (HA) in NIH/3T3 cells stably expressing TaPIN1 and NIH/3T3 cells stably expressing hPIN1; counterstaining with DAPI. All the results are representatives of three independent experiments. Objective used, 60×.
Extended Data Figure 5 TaPIN1 functionally replaces hPIN1.
a, The HA-tagged version of TaPIN1 and hPIN1 can rescue cell spreading defects in Pin1−/− knockout murine immortalized fibroblasts. Cell spreading was assessed by Phalloidin-TRITC staining. Con, control (transfection with the appropriate empty vector). b, TaPIN1 causes centrosome amplification. NIH/3T3 fibroblasts stably expressing TaPIN1 or hPIN1 were arrested at the G1/S transition by aphidicolin, stained with anti-γ-tubulin antibody (arrow) and counterstained with DAPI. Photographs are representative of one cell in three independent experiments. Objective used, 60×.
Extended Data Figure 6 The PPIase domain of PIN1 is well conserved.
a, Sequence alignment of PIN1 genes in H. sapiens, A. thaliana, T. brucei and T. annulata revealed the presence or the absence of a conserved WW domain. Percentages of identity are indicated. The magenta box indicates the predicted signal peptide of TaPIN1. b, Homology models for TaPIN1 wild type (WT) and the A53P mutant based on sequence identity/similarity with hPIN1. The Ala to Pro mutation in TaPIN1 appears to induce a conformational change within and nearby the catalytic loop. c, The buparvaquone molecule can be docked in the active site of hPIN1 or in the active site of the wild-type TaPIN1 predicted structure. Here, the second lowest docked energy pose is shown (the best predicted energy pose is shown Fig. 3a). These two predicted binding poses are fully consistent with the different binding modes of some inhibitors co-crystallized with hPIN1. Yet, considering the computations shown in e, we suggest that the most likely pose for buparvaquone corresponds to pose 1, as reported in Fig. 3a. However, independently of the selected poses, we expect that buparvaquone would not fit well in the catalytic pocket due to structural changes induced by the A53P mutation. d, Three-dimensional structure of the experimental hPIN1 structure and the predicted TaPIN1 wild-type and A53P mutant structures, alongside Trypanosome TbPIN and Arabidopsis PIN1At models (ribbon diagram). The three-dimensional structures are well conserved among these proteins with some differences, for instance, in the catalytic loops. The TaPIN1, TbPIN1 and PIN1At enzymes lack the WW domain present in hPIN1 yet the overall fold in the catalytic area is well conserved, suggesting that accurate homology models for TaPIN1 can be built using the approach used here. e, The experimental structure of hPIN1 is represented as a solid surface with a view down the active site, showing a small co-crystallized ligand next to the catalytic site residue Cys 113. In the same orientation, small chemical fragments are predicted to bind in the catalytic site region with FTmap. Using this information, we propose that the most likely binding pose for buparvaquone is the one shown in Fig. 3.
Extended Data Figure 7 Buparvaquone-resistant parasites are mutated in the TaPIN1 PPIase domain.
a, Sequence alignment of the Tapin1 gene in T. annulata wild-type (WT) or buparvaquone-resistant genomes (NCBI BLAST). The annotated T. annulata intron is indicated in grey. Buparvaquone-resistant parasites have three mutations indicated in black. b, Sequence alignment of the TaPIN1 protein in T. annulata wild-type or buparvaquone-resistant strains. Buparvaquone-resistant parasites have a mutation in the PPIase domain (residue 53): Ala to Pro mutation. c, Signal peptide predicted of TaPIN1(A53P) with SignaIP 3.0 Server. d, The measure of TaPIN1–GST and TaPIN1(A53P)–GST catalytic activity were determined using PPIase assay (chymotrypsin-coupled assay) (average ± s.d., n = 4).
Extended Data Figure 8 Inhibition of BtPin1 does not affect Theileria-associated cell transformation.
a, Buparvaquone (Bup), juglone (Jug) and DTM decreased the viability of cells infected with T. parva (TpMD409). Host cell viability was assessed using the XTT assay after 72 h. b, Colony formation of parasite-infected TpMD409 cells in soft agar was lost after 72 h treatment with buparvaquone, juglone or DTM. Numbers of macroscopic colonies per plate were counted after 10 days. c, siBtPin1 has no effect on TBL3 transformed phenotypes. TBL3 cells were transiently transfected with siControl or siBtPin1. The average number of colonies per plate is shown (average ± s.d., n = 3). Indicated protein levels were detected by western blot analysis using specific antibodies. Actin was used as loading control (results are representative of three independent experiments). d–f, Overexpression of the TaPIN1 A53P mutant rescued the buparvaquone but not juglone effects on Theileria-infected cells. WT, wild type. All data represent three independent experiments (average ± s.d., n = 3). The SPSS 19.0 program was used for statistics. P < 0.05, P < 0.01. NS, not significant.
Extended Data Figure 9 TaPIN1 changes c-JUN stability through the regulation of FBW7.
a, TaPIN1 interacts with endogenous mouse FBW7. Protein extracts from NIH/3T3 cells stably expressing HA–Flag–TaPIN1 or HA–Flag–Control (Con) were used for HA immunoprecipitation, followed by immunoblot analysis using FBW7 and HA antibodies. b, TaPIN1 interacts with endogenous bovine FBW7 protein. Parasite-infected TBL3 and TpMD409 cell lysates were incubated with the indicated GST-coupled beads, followed by immunoblot analysis using a FBW7 antibody. c, d, Analysis of indicated bovine gene expression by qPCR in TBL3 cells following buparvaquone (Bup) or juglone (Jug) treatment with or without TaPIN1 wild type (WT) or A53P mutant. β-Actin and H2A mRNAs were used for normalization (data represent three independent experiments; average ± s.d., n = 3). e, siBtPin1 does not affect FBW7α or c-JUN protein levels. TBL3 were transiently transfected by siControl (siCon) or siBtPin1. Indicated protein levels were detected by western blot analysis using specific antibodies. Actin was used as loading control. f, Inhibition of TaPIN1 by buparvaquone/juglone increased c-JUN ubiquitination and decreased FBW7 ubiquitination. Parasite-infected TBL3 cells (± buparvaquone or juglone) were incubated with MG132, followed by immunoprecipitation of endogenous c-JUN or FBW7 and immunoblot analysis with the indicated antibodies. Con, control. g, The effect of buparvaquone on c-JUN ubiquitination was rescued by overexpression of TaPIN1(A53P). Parasite-infected TBL3 cells (± buparvaquone or juglone) were transfected with TaPIN1 wild type, TaPIN1(A53P) or empty vector (Con) and then treated with MG132, followed by immunoprecipitation of endogenous c-JUN and immunoblot analysis with the indicated antibodies. h, Measure of c-MYC, KLF5 and activated NOTCH1 protein levels in TBL3 cells upon buparvaquone treatment. Tubulin was used as loading control. i, Measure of c-MYC, KLF5 and activated NOTCH1 protein levels in TBL3 cells upon FBW7α ectopic expression. Con, control (transfection with the appropriate empty vector). Tubulin was used as loading control. a, b, e–h, All the results are representative of three independent experiments. NS, not significant.
Extended Data Figure 10 Original blot.
a, From Fig. 1b: TaPIN1 protein was detected in the host cytoplasm and nucleus, in contrast with apicomplexan actin (TaActin). Bovine histone H3 (nuclear) and tubulin (cytoplasmic) proteins were controls. b, From Fig. 4a: endogenous TaPIN1 interacts with FBW7α isoform. Protein extracts from TBL3 expressing Flag–hFBW7 isoforms or Flag–control (Con) were immunoprecipitated and immunoblotted with TaPIN1 or Flag antibodies. c, From Fig. 4b: inhibition of TaPIN1 by buparvaquone (Bup), juglone (Jug) or DTM increased FBW7α protein levels and decreased c-JUN expression in TBL3 cells. Actin/tubulin were loading controls. d, From Fig. 4c: Inhibition of Fbw7 increases c-JUN protein levels in TBL3 cells, whereas ectopic FBW7α expression reduced c-JUN protein levels. Bovine actin/tubulin were loading controls. Con, empty vector. e, From Fig. 4h: efficiency of two independent c-Jun-targeting siRNAs. Bovine tubulin loading control.
Rights and permissions
About this article
Cite this article
Marsolier, J., Perichon, M., DeBarry, J. et al. Theileria parasites secrete a prolyl isomerase to maintain host leukocyte transformation. Nature 520, 378–382 (2015). https://doi.org/10.1038/nature14044
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14044
This article is cited by
-
Cancerogenic parasites in veterinary medicine: a narrative literature review
Infectious Agents and Cancer (2023)
-
Dual RNA-seq to catalogue host and parasite gene expression changes associated with virulence of T. annulata-transformed bovine leukocytes: towards identification of attenuation biomarkers
Scientific Reports (2023)
-
Theileria annulata SVSP455 interacts with host HSP60
Parasites & Vectors (2022)
-
Metabolomic profiling of bovine leucocytes transformed by Theileria annulata under BW720c treatment
Parasites & Vectors (2022)
-
Trifloxystrobin blocks the growth of Theileria parasites and is a promising drug to treat Buparvaquone resistance
Communications Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.